Loss of the p53/p63 regulated desmosomal protein Perp promotes tumorigenesis
- PMID: 20975948
- PMCID: PMC2958815
- DOI: 10.1371/journal.pgen.1001168
Loss of the p53/p63 regulated desmosomal protein Perp promotes tumorigenesis
Abstract
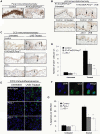
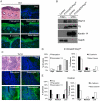
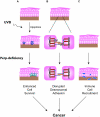
Dysregulated cell-cell adhesion plays a critical role in epithelial cancer development. Studies of human and mouse cancers have indicated that loss of adhesion complexes known as adherens junctions contributes to tumor progression and metastasis. In contrast, little is known regarding the role of the related cell-cell adhesion junction, the desmosome, during cancer development. Studies analyzing expression of desmosome components during human cancer progression have yielded conflicting results, and therefore genetic studies using knockout mice to examine the functional consequence of desmosome inactivation for tumorigenesis are essential for elucidating the role of desmosomes in cancer development. Here, we investigate the consequences of desmosome loss for carcinogenesis by analyzing conditional knockout mice lacking Perp, a p53/p63 regulated gene that encodes an important component of desmosomes. Analysis of Perp-deficient mice in a UVB-induced squamous cell skin carcinoma model reveals that Perp ablation promotes both tumor initiation and progression. Tumor development is associated with inactivation of both of Perp's known functions, in apoptosis and cell-cell adhesion. Interestingly, Perp-deficient tumors exhibit widespread downregulation of desmosomal constituents while adherens junctions remain intact, suggesting that desmosome loss is a specific event important for tumorigenesis rather than a reflection of a general change in differentiation status. Similarly, human squamous cell carcinomas display loss of PERP expression with retention of adherens junctions components, indicating that this is a relevant stage of human cancer development. Using gene expression profiling, we show further that Perp loss induces a set of inflammation-related genes that could stimulate tumorigenesis. Together, these studies suggest that Perp-deficiency promotes cancer by enhancing cell survival, desmosome loss, and inflammation, and they highlight a fundamental role for Perp and desmosomes in tumor suppression. An understanding of the factors affecting cancer progression is important for ultimately improving the diagnosis, prognostication, and treatment of cancer.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Cooper GM. Oncogenes. Boston: Jones and Bartlett Publishers; 1995. p. xv, 384.
-
- Alam M, Ratner D. Cutaneous squamous-cell carcinoma. N Engl J Med. 2001;344:975–983. - PubMed
-
- Hunter KD, Parkinson EK, Harrison PR. Profiling early head and neck cancer. Nat Rev Cancer. 2005;5:127–135. - PubMed
-
- Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nat Rev Genet. 2002;3:199–209. - PubMed
-
- Green KJ, Gaudry CA. Are desmosomes more than tethers for intermediate filaments? Nat Rev Mol Cell Biol. 2000;1:208–216. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

