Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis
- PMID: 20976044
- PMCID: PMC2957403
- DOI: 10.1371/journal.pbio.1000520
Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis
Abstract
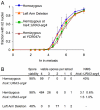
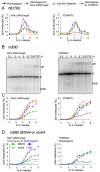
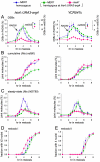
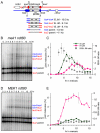
Recombination between homologous chromosomes of different parental origin (homologs) is necessary for their accurate segregation during meiosis. It has been suggested that meiotic inter-homolog recombination is promoted by a barrier to inter-sister-chromatid recombination, imposed by meiosis-specific components of the chromosome axis. Consistent with this, measures of Holliday junction-containing recombination intermediates (joint molecules [JMs]) show a strong bias towards inter-homolog and against inter-sister JMs. However, recombination between sister chromatids also has an important role in meiosis. The genomes of diploid organisms in natural populations are highly polymorphic for insertions and deletions, and meiotic double-strand breaks (DSBs) that form within such polymorphic regions must be repaired by inter-sister recombination. Efforts to study inter-sister recombination during meiosis, in particular to determine recombination frequencies and mechanisms, have been constrained by the inability to monitor the products of inter-sister recombination. We present here molecular-level studies of inter-sister recombination during budding yeast meiosis. We examined events initiated by DSBs in regions that lack corresponding sequences on the homolog, and show that these DSBs are efficiently repaired by inter-sister recombination. This occurs with the same timing as inter-homolog recombination, but with reduced (2- to 3-fold) yields of JMs. Loss of the meiotic-chromosome-axis-associated kinase Mek1 accelerates inter-sister DSB repair and markedly increases inter-sister JM frequencies. Furthermore, inter-sister JMs formed in mek1Δ mutants are preferentially lost, while inter-homolog JMs are maintained. These findings indicate that inter-sister recombination occurs frequently during budding yeast meiosis, with the possibility that up to one-third of all recombination events occur between sister chromatids. We suggest that a Mek1-dependent reduction in the rate of inter-sister repair, combined with the destabilization of inter-sister JMs, promotes inter-homolog recombination while retaining the capacity for inter-sister recombination when inter-homolog recombination is not possible.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet. 2007;16 Spec No. 2:R203–R208. - PubMed
-
- Keeney S, Giroux C. N, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. - PubMed
-
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

