A kernelisation approach for multiple d-Hitting Set and its application in optimal multi-drug therapeutic combinations
- PMID: 20976188
- PMCID: PMC2956629
- DOI: 10.1371/journal.pone.0013055
A kernelisation approach for multiple d-Hitting Set and its application in optimal multi-drug therapeutic combinations
Abstract
Therapies consisting of a combination of agents are an attractive proposition, especially in the context of diseases such as cancer, which can manifest with a variety of tumor types in a single case. However uncovering usable drug combinations is expensive both financially and temporally. By employing computational methods to identify candidate combinations with a greater likelihood of success we can avoid these problems, even when the amount of data is prohibitively large. Hitting Set is a combinatorial problem that has useful application across many fields, however as it is NP-complete it is traditionally considered hard to solve exactly. We introduce a more general version of the problem (α,β,d)-Hitting Set, which allows more precise control over how and what the hitting set targets. Employing the framework of Parameterized Complexity we show that despite being NP-complete, the (α,β,d)-Hitting Set problem is fixed-parameter tractable with a kernel of size O(αdk(d)) when we parameterize by the size k of the hitting set and the maximum number α of the minimum number of hits, and taking the maximum degree d of the target sets as a constant. We demonstrate the application of this problem to multiple drug selection for cancer therapy, showing the flexibility of the problem in tailoring such drug sets. The fixed-parameter tractability result indicates that for low values of the parameters the problem can be solved quickly using exact methods. We also demonstrate that the problem is indeed practical, with computation times on the order of 5 seconds, as compared to previous Hitting Set applications using the same dataset which exhibited times on the order of 1 day, even with relatively relaxed notions for what constitutes a low value for the parameters. Furthermore the existence of a kernelization for (α,β,d)-Hitting Set indicates that the problem is readily scalable to large datasets.
Conflict of interest statement
Figures
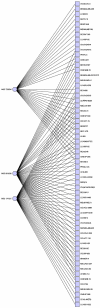







 to
to  . This hitting set also maximizes the number of hits on
the melanoma cell lines. Only cell lines with at least one adjacent
compound are shown.
. This hitting set also maximizes the number of hits on
the melanoma cell lines. Only cell lines with at least one adjacent
compound are shown.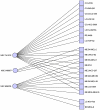
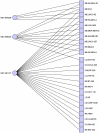
Similar articles
-
Optimal drug combinations and minimal hitting sets.BMC Syst Biol. 2009 Aug 6;3:81. doi: 10.1186/1752-0509-3-81. BMC Syst Biol. 2009. PMID: 19660129 Free PMC article.
-
Fixed-parameter tractability of the maximum agreement supertree problem.IEEE/ACM Trans Comput Biol Bioinform. 2010 Apr-Jun;7(2):342-53. doi: 10.1109/TCBB.2008.93. IEEE/ACM Trans Comput Biol Bioinform. 2010. PMID: 20431153
-
Parameterized complexity analysis in computational biology.Comput Appl Biosci. 1995 Feb;11(1):49-57. doi: 10.1093/bioinformatics/11.1.49. Comput Appl Biosci. 1995. PMID: 7796275
-
Combining targeted therapies: practical issues to consider at the bench and bedside.Oncologist. 2010;15(1):37-50. doi: 10.1634/theoncologist.2009-0117. Epub 2010 Jan 15. Oncologist. 2010. PMID: 20080862 Free PMC article. Review.
-
Predictive approaches for drug combination discovery in cancer.Brief Bioinform. 2018 Mar 1;19(2):263-276. doi: 10.1093/bib/bbw104. Brief Bioinform. 2018. PMID: 27881431 Free PMC article. Review.
Cited by
-
Systematic quantitative characterization of cellular responses induced by multiple signals.BMC Syst Biol. 2011 May 30;5:88. doi: 10.1186/1752-0509-5-88. BMC Syst Biol. 2011. PMID: 21624115 Free PMC article.
-
Branched-Chain Amino Acids Catabolism Pathway Regulation Plays a Critical Role in the Improvement of Leukopenia Induced by Cyclophosphamide in 4T1 Tumor-Bearing Mice Treated With Lvjiaobuxue Granule.Front Pharmacol. 2021 Oct 25;12:657047. doi: 10.3389/fphar.2021.657047. eCollection 2021. Front Pharmacol. 2021. PMID: 34759816 Free PMC article.
-
The landscape of receptor-mediated precision cancer combination therapy via a single-cell perspective.Nat Commun. 2022 Mar 25;13(1):1613. doi: 10.1038/s41467-022-29154-2. Nat Commun. 2022. PMID: 35338126 Free PMC article.
References
-
- Albain KS, Crowley JJ, LeBlanc M, Livingston RB. Determinants of improved outcome in small-cell lung cancer: an analysis of the 2,580-patient southwest oncology group data base. Journal of Clinical Oncology. 1990;8:1563–1574. - PubMed
-
- Flamant F, Schwartz L, Delons E, Caillaud JM, Hartmann O, et al. Nonseminomatous malignant germ cell tumors in children. Multidrug therapy in stages III and IV. Cancer. 1984;54:1687–1691. - PubMed
-
- Fu KK, Silverberg IJ, Phillips TL, Friedman MA. Combined radiotherapy and multidrug chemotherapy for advanced head and neck cancer: results of a radiation therapy oncology group pilot study. Cancer Treatment Reports. 1979;63:351–357. - PubMed
-
- Berman P, DasGupta B, Sontag ED. Randomized approximation algorithms for set multicover problems with applications to reverse engineering of protein and gene networks. Discrete Applied Mathematics. 2007;155:733–749.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

