Epstein Barr virus-encoded EBNA1 interference with MHC class I antigen presentation reveals a close correlation between mRNA translation initiation and antigen presentation
- PMID: 20976201
- PMCID: PMC2954899
- DOI: 10.1371/journal.ppat.1001151
Epstein Barr virus-encoded EBNA1 interference with MHC class I antigen presentation reveals a close correlation between mRNA translation initiation and antigen presentation
Abstract
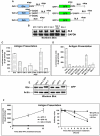
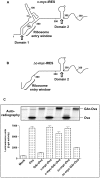
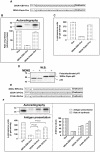
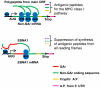
Viruses are known to employ different strategies to manipulate the major histocompatibility (MHC) class I antigen presentation pathway to avoid recognition of the infected host cell by the immune system. However, viral control of antigen presentation via the processes that supply and select antigenic peptide precursors is yet relatively unknown. The Epstein-Barr virus (EBV)-encoded EBNA1 is expressed in all EBV-infected cells, but the immune system fails to detect and destroy EBV-carrying host cells. This immune evasion has been attributed to the capacity of a Gly-Ala repeat (GAr) within EBNA1 to inhibit MHC class I restricted antigen presentation. Here we demonstrate that suppression of mRNA translation initiation by the GAr in cis is sufficient and necessary to prevent presentation of antigenic peptides from mRNAs to which it is fused. Furthermore, we demonstrate a direct correlation between the rate of translation initiation and MHC class I antigen presentation from a certain mRNA. These results support the idea that mRNAs, and not the encoded full length proteins, are used for MHC class I restricted immune surveillance. This offers an additional view on the role of virus-mediated control of mRNA translation initiation and of the mechanisms that control MHC class I restricted antigen presentation in general.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. - PubMed
-
- Rock KL, York IA, Goldberg AL. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat Immunol. 2004;5:670–677. - PubMed
-
- Kloetzel PM. Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat Immunol. 2004;5:661–669. - PubMed
-
- Van Kaer L, Ashton-Rickardt PG, Ploegh HL, Tonegawa S. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4-8+ T cells. Cell. 1992;71:1205–1214. - PubMed
-
- Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

