Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat
- PMID: 20976203
- PMCID: PMC2954905
- DOI: 10.1371/journal.ppat.1001152
Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat
Abstract
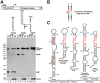
The human immunodeficiency virus 1 (HIV-1) transcriptional transactivator (Tat) is essential for synthesis of full-length transcripts from the integrated viral genome by RNA polymerase II (Pol II). Tat recruits the host positive transcription elongation factor b (P-TEFb) to the HIV-1 promoter through binding to the transactivator RNA (TAR) at the 5'-end of the nascent HIV transcript. P-TEFb is a general Pol II transcription factor; its cellular activity is controlled by the 7SK small nuclear RNA (snRNA) and the HEXIM1 protein, which sequester P-TEFb into transcriptionally inactive 7SK/HEXIM/P-TEFb snRNP. Besides targeting P-TEFb to HIV transcription, Tat also increases the nuclear level of active P-TEFb through promoting its dissociation from the 7SK/HEXIM/P-TEFb RNP by an unclear mechanism. In this study, by using in vitro and in vivo RNA-protein binding assays, we demonstrate that HIV-1 Tat binds with high specificity and efficiency to an evolutionarily highly conserved stem-bulge-stem motif of the 5'-hairpin of human 7SK snRNA. The newly discovered Tat-binding motif of 7SK is structurally and functionally indistinguishable from the extensively characterized Tat-binding site of HIV TAR and importantly, it is imbedded in the HEXIM-binding elements of 7SK snRNA. We show that Tat efficiently replaces HEXIM1 on the 7SK snRNA in vivo and therefore, it promotes the disassembly of the 7SK/HEXIM/P-TEFb negative transcriptional regulatory snRNP to augment the nuclear level of active P-TEFb. This is the first demonstration that HIV-1 specifically targets an important cellular regulatory RNA, most probably to promote viral transcription and replication. Demonstration that the human 7SK snRNA carries a TAR RNA-like Tat-binding element that is essential for the normal transcriptional regulatory function of 7SK questions the viability of HIV therapeutic approaches based on small drugs blocking the Tat-binding site of HIV TAR.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



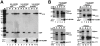

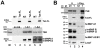
Similar articles
-
A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb.Mol Cell Biol. 2004 Jun;24(12):5094-105. doi: 10.1128/MCB.24.12.5094-5105.2004. Mol Cell Biol. 2004. PMID: 15169877 Free PMC article.
-
The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK.PLoS One. 2010 Aug 23;5(8):e12335. doi: 10.1371/journal.pone.0012335. PLoS One. 2010. PMID: 20808803 Free PMC article.
-
Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding.Mol Cell Biol. 2006 Jan;26(2):630-42. doi: 10.1128/MCB.26.2.630-642.2006. Mol Cell Biol. 2006. PMID: 16382153 Free PMC article.
-
New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC).J Neuroimmune Pharmacol. 2011 Jun;6(2):260-8. doi: 10.1007/s11481-011-9267-6. Epub 2011 Mar 1. J Neuroimmune Pharmacol. 2011. PMID: 21360054 Free PMC article. Review.
-
[Misregulation of P-TEFb activity: pathological consequences].Med Sci (Paris). 2012 Feb;28(2):200-5. doi: 10.1051/medsci/2012282019. Epub 2012 Feb 27. Med Sci (Paris). 2012. PMID: 22377309 Review. French.
Cited by
-
Structural transitions in the RNA 7SK 5' hairpin and their effect on HEXIM binding.Nucleic Acids Res. 2020 Jan 10;48(1):373-389. doi: 10.1093/nar/gkz1071. Nucleic Acids Res. 2020. PMID: 31732748 Free PMC article.
-
A Natural Product from Polygonum cuspidatum Sieb. Et Zucc. Promotes Tat-Dependent HIV Latency Reversal through Triggering P-TEFb's Release from 7SK snRNP.PLoS One. 2015 Nov 16;10(11):e0142739. doi: 10.1371/journal.pone.0142739. eCollection 2015. PLoS One. 2015. PMID: 26569506 Free PMC article.
-
The RNA-Binding Proteins SRP14 and HMGB3 Control HIV-1 Tat mRNA Processing and Translation During HIV-1 Latency.Front Genet. 2021 Jun 14;12:680725. doi: 10.3389/fgene.2021.680725. eCollection 2021. Front Genet. 2021. PMID: 34194479 Free PMC article.
-
Role of Host Factors on the Regulation of Tat-Mediated HIV-1 Transcription.Curr Pharm Des. 2017;23(28):4079-4090. doi: 10.2174/1381612823666170622104355. Curr Pharm Des. 2017. PMID: 28641539 Free PMC article. Review.
-
Depicting HIV-1 Transcriptional Mechanisms: A Summary of What We Know.Viruses. 2020 Dec 3;12(12):1385. doi: 10.3390/v12121385. Viruses. 2020. PMID: 33287435 Free PMC article. Review.
References
-
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. - PubMed
-
- Nechaev S, Adelman K. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle. 2008;7:1539–1544. - PubMed
-
- Price DH. Poised polymerases: on your mark…get set…go! Mol Cell. 2008;30:7–10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

