Evolutionary mode and functional divergence of vertebrate NMDA receptor subunit 2 genes
- PMID: 20976280
- PMCID: PMC2954789
- DOI: 10.1371/journal.pone.0013342
Evolutionary mode and functional divergence of vertebrate NMDA receptor subunit 2 genes
Abstract
Background: Ionotropic glutamate receptors in the central nervous system play a major role in numerous brain functions including learning and memory in many vertebrate species. NR2 subunits have been regarded as rate-limiting molecules in controlling the optimal N-methyl-D-aspartate (NMDA) receptor's coincidence-detection property and subsequent learning and memory function across multi-species. However, its evolutionary mode among vertebrate species remains unclear.
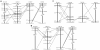
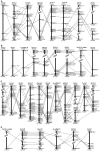
Results: With extensive analysis of phylogeny, exon structure, protein domain, paralogon and synteny, we demonstrated that two-round genome duplication generated quartet GRIN2 genes and the third-round fish-specific genome duplication generated extra copies of fish GRIN2 genes. In addition, in-depth investigation has enabled the identification of three novel genes, GRIN2C_Gg, GRIN2D-1_Ol and GRIN2D-2_Tr in the chicken, medaka and fugu genome, respectively. Furthermore, we showed functional divergence of NR2 genes mostly occurred at the first-round duplication, amino acid residues located at the N-terminal Lig_chan domain were responsible for type I functional divergence between these GRIN2 subfamilies and purifying selection has been the prominent natural pressure operating on these diversified GRIN2 genes.
Conclusion and significance: These findings provide intriguing subjects for testing the 2R and 3R hypothesis and we expect it could provide new insights into the underlying evolution mechanisms of cognition in vertebrate.
Conflict of interest statement
Figures



Similar articles
-
Evolution of NMDA receptor cytoplasmic interaction domains: implications for organisation of synaptic signalling complexes.BMC Neurosci. 2008 Jan 15;9:6. doi: 10.1186/1471-2202-9-6. BMC Neurosci. 2008. PMID: 18197970 Free PMC article.
-
Evolution of vertebrate nicotinic acetylcholine receptors.BMC Evol Biol. 2019 Jan 30;19(1):38. doi: 10.1186/s12862-018-1341-8. BMC Evol Biol. 2019. PMID: 30700248 Free PMC article.
-
OHNOLOGS v2: a comprehensive resource for the genes retained from whole genome duplication in vertebrates.Nucleic Acids Res. 2020 Jan 8;48(D1):D724-D730. doi: 10.1093/nar/gkz909. Nucleic Acids Res. 2020. PMID: 31612943 Free PMC article.
-
New Insights Into the Evolutionary History of Melatonin Receptors in Vertebrates, With Particular Focus on Teleosts.Front Endocrinol (Lausanne). 2020 Sep 24;11:538196. doi: 10.3389/fendo.2020.538196. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 33071966 Free PMC article. Review.
-
Evolution of endothelin receptors in vertebrates.Gen Comp Endocrinol. 2014 Dec 1;209:21-34. doi: 10.1016/j.ygcen.2014.06.028. Epub 2014 Jul 8. Gen Comp Endocrinol. 2014. PMID: 25010382 Review.
Cited by
-
Detection of Regional Variation in Selection Intensity within Protein-Coding Genes Using DNA Sequence Polymorphism and Divergence.Mol Biol Evol. 2017 Nov 1;34(11):3006-3022. doi: 10.1093/molbev/msx213. Mol Biol Evol. 2017. PMID: 28962009 Free PMC article.
-
Phylogenetic analysis of ionotropic L-glutamate receptor genes in the Bilateria, with special notes on Aplysia californica.BMC Evol Biol. 2017 Jan 11;17(1):11. doi: 10.1186/s12862-016-0871-1. BMC Evol Biol. 2017. PMID: 28077092 Free PMC article.
-
Evolution, functional divergence and conserved exon-intron structure of bHLH/PAS gene family.Mol Genet Genomics. 2014 Feb;289(1):25-36. doi: 10.1007/s00438-013-0786-0. Epub 2013 Nov 8. Mol Genet Genomics. 2014. PMID: 24202550
-
Genetic variation of rs12918566 affects GRIN2A expression and is associated with spontaneous movement response during sevoflurane anesthesia induction.Brain Behav. 2021 Aug;11(8):e02165. doi: 10.1002/brb3.2165. Epub 2021 Jul 21. Brain Behav. 2021. PMID: 34291608 Free PMC article.
-
Glutamate and ATP at the Interface Between Signaling and Metabolism in Astroglia: Examples from Pathology.Neurochem Res. 2017 Jan;42(1):19-34. doi: 10.1007/s11064-016-1848-6. Epub 2016 Feb 25. Neurochem Res. 2017. PMID: 26915104 Review.
References
-
- Burrell BD, Sahley CL. Learning in simple systems. Curr Opin Neurobiol. 2001;11:757–764. - PubMed
-
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. - PubMed
-
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. - PubMed
-
- Tsien JZ, Huerta PT, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87:1327–1338. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

