Delayed c-Fos activation in human cells triggers XPF induction and an adaptive response to UVC-induced DNA damage and cytotoxicity
- PMID: 20976523
- PMCID: PMC3078315
- DOI: 10.1007/s00018-010-0546-9
Delayed c-Fos activation in human cells triggers XPF induction and an adaptive response to UVC-induced DNA damage and cytotoxicity
Abstract
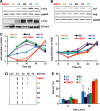
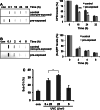
The oncoprotein c-Fos has been commonly found differently expressed in cancer cells. Our previous work showed that mouse cells lacking the immediate-early gene c-fos are hypersensitive to ultraviolet (UVC) light. Here, we demonstrate that in human diploid fibroblasts UV-triggered induction of c-Fos protein is a delayed and long-lasting event. Sustained upregulation of c-Fos goes along with transcriptional stimulation of the NER gene xpf, which harbors an AP-1 binding site in the promoter. Data gained on c-Fos knockdown and c-Fos overexpressing human cells provide evidence that c-Fos/AP-1 stimulates upregulation of XPF, thereby increasing the cellular repair capacity protecting from UVC-induced DNA damage. When these cells are pre-exposed to a low non-toxic UVC dose and challenged with a subsequent high dose of UVC irradiation, they show accelerated repair of UVC-induced DNA adducts and reduced cell kill. The data indicate a protective role of c-Fos induction by triggering an adaptive response pathway.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures






References
-
- Christmann M, Fritz G, Kaina B (2007) Induction of DNA repair genes in mammalian cells in response to genotoxic stress. In: D. Lankenau (Ed) Genome dynamics and stability, vol 1. Springer, Berlin, pp 383–398
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

