Participation of CYP2C8 and CYP3A4 in the N-demethylation of imatinib in human hepatic microsomes
- PMID: 20977456
- PMCID: PMC2998687
- DOI: 10.1111/j.1476-5381.2010.00946.x
Participation of CYP2C8 and CYP3A4 in the N-demethylation of imatinib in human hepatic microsomes
Abstract
Background and purpose: Imatinib is a clinically important inhibitor of tyrosine kinases that are dysregulated in chronic myelogenous leukaemia and gastrointestinal stromal tumours. Inter-individual variation in imatinib pharmacokinetics is extensive, and influences drug safety and efficacy. Hepatic cytochrome P450 (CYP) 3A4 has been implicated in imatinib N-demethylation, but the clearance of imatinib decreases during prolonged therapy. CYP3A phenotype correlates with imatinib clearance at the commencement of therapy, but not at steady state. The present study evaluated the possibility that multiple CYPs may contribute to imatinib oxidation in liver.
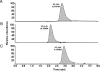
Experimental approach: Imatinib biotransformation in human liver microsomes (n= 20) and by cDNA-expressed CYPs was determined by LC-MS. Relationships between imatinib N-demethylation and other drug metabolizing CYPs were assessed.
Key results: N-desmethylimatinib formation was correlated with microsomal oxidation of the CYP3A4 substrates testosterone (ρ= 0.60; P < 0.01) and midazolam (ρ= 0.46; P < 0.05), and the CYP2C8 substrate paclitaxel (ρ= 0.58; P < 0.01). cDNA-derived CYPs 2C8, 3A4, 3A5 and 3A7 supported imatinib N-demethylation, but 10 other CYPs were inactive; in kinetic studies, CYP2C8 was a high-affinity enzyme with a catalytic efficiency ∼15-fold greater than those of CYPs 3A4 and 3A5. The CYP3A inhibitors ketoconazole and troleandomycin, and the CYP2C8 inhibitors quercetin and paclitaxel decreased imatinib oxidation. From molecular modelling, the imatinib structure could be superimposed on a pharmacophore for CYP2C8 substrates.
Conclusions and implications: CYP2C8 and CYPs 3A contribute to imatinib N-demethylation in human liver. The involvement of CYP2C8 may account in part for the wide inter-patient variation in imatinib pharmacokinetics observed in clinical practice.
© 2010 The Authors. British Journal of Pharmacology © 2010 The British Pharmacological Society.
Figures

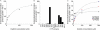

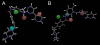
References
-
- Apperley JF. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8:1018–1029. - PubMed
-
- Ayoub WS, Geller SA, Tran T, Martin P, Vierling JM, Poordad FF. Imatinib (Gleevec)-induced hepatotoxicity. J Clin Gastroenterol. 2005;39:75–77. - PubMed
-
- Bahadur N, Leathart JB, Mutch E, Steimel-Crespi D, Dunn SA, Gilissen R, et al. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6α-hydroxylase activity in human liver microsomes. Biochem Pharmacol. 2002;64:1579–1589. - PubMed
-
- Blasdel C, Egorin MJ, Lagattuta TF, Druker BJ, Deininger MW. Therapeutic drug monitoring in CML patients on imatinib. Blood. 2007;110:1699–1701. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

