ZEB1-responsive genes in non-small cell lung cancer
- PMID: 20980099
- PMCID: PMC3337721
- DOI: 10.1016/j.canlet.2010.09.007
ZEB1-responsive genes in non-small cell lung cancer
Abstract
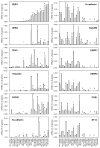
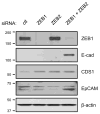
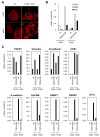
The epithelial to mesenchymal transition (EMT) is a developmental process enabling epithelial cells to gain a migratory mesenchymal phenotype. In cancer, this process contributes to metastases; however the regulatory signals and mechanistic details are not fully elucidated. Here, we sought to identify the subset of genes regulated in lung cancer by ZEB1, an E-box transcriptional repressor known to induce EMT. Using an Affymetrix-based expression database of 38 non-small cell lung cancer (NSCLC) cell lines, we identified 324 genes that correlated negatively with ZEB1 and 142 that were positively correlated. A mesenchymal gene pattern (low E-cadherin, high Vimentin or N-cadherin) was significantly associated with ZEB1 and ZEB2, but not with Snail, Slug, Twist1 or Twist2. Among eight genes selected for validation, seven were confirmed to correlate with ZEB1 by quantitative real-time RT-PCR in a series of 22 NSCLC cell lines, either negatively (CDS1, EpCAM, ESRP1, ESRP2, ST14) or positively (FGFR1, Vimentin). In addition, over-expression or knockdown of ZEB1 led to corresponding changes in gene expression, demonstrating that these genes are also regulated by ZEB1, either directly or indirectly. Of note, the combined knockdown of ZEB1 and ZEB2 led to apparent synergistic responses in gene expression. Furthermore, these responses were not restricted to artificial settings, since most genes were similarly regulated during a physiologic induction of EMT by TGF-β plus EGF. Finally, the absence of ST14 (matriptase) was linked to ZEB1 positivity in lung cancer tissue microarrays, implying that the regulation observed in vitro applies to the human disease. In summary, this study identifies a new set of ZEB-regulated genes in human lung cancer cells and supports the hypothesis that ZEB1 and ZEB2 are key regulators of the EMT process in this disease.
Copyright © 2010 Elsevier Ireland Ltd. All rights reserved.
Conflict of interest statement
We have no conflict of interest to de disclosed.
Figures



References
-
- Marugame T, Yoshimi I. Comparison of cancer mortality (lung cancer) in five countries: France, Italy, Japan, UK and USA from the WHO Mortality Database (1960–2000) Jpn J Clin Oncol. 2005;35:168–70. - PubMed
-
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. - PubMed
-
- Sabbah M, Emami S, Redeuilh G, Julien S, Prevost G, Zimber A, Ouelaa R, Bracke M, De Wever O, Gespach C. Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers. Drug Resist Updat. 2008;11:123–51. - PubMed
-
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. - PubMed
-
- Voulgari A, Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta. 2009;1796:75–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

