STAT1 is a master regulator of pancreatic {beta}-cell apoptosis and islet inflammation
- PMID: 20980260
- PMCID: PMC3020778
- DOI: 10.1074/jbc.M110.162131
STAT1 is a master regulator of pancreatic {beta}-cell apoptosis and islet inflammation
Abstract
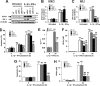
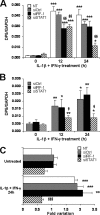
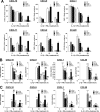
Cytokines produced by islet-infiltrating immune cells induce β-cell apoptosis in type 1 diabetes. The IFN-γ-regulated transcription factors STAT1/IRF-1 have apparently divergent effects on β-cells. Thus, STAT1 promotes apoptosis and inflammation, whereas IRF-1 down-regulates inflammatory mediators. To understand the molecular basis for these differential outcomes within a single signal transduction pathway, we presently characterized the gene networks regulated by STAT1 and IRF-1 in β-cells. This was done by using siRNA approaches coupled to microarray analysis of insulin-producing cells exposed or not to IL-1β and IFN-γ. Relevant microarray findings were further studied in INS-1E cells and primary rat β-cells. STAT1, but not IRF-1, mediates the cytokine-induced loss of the differentiated β-cell phenotype, as indicated by decreased insulin, Pdx1, MafA, and Glut2. Furthermore, STAT1 regulates cytokine-induced apoptosis via up-regulation of the proapoptotic protein DP5. STAT1 and IRF-1 have opposite effects on cytokine-induced chemokine production, with IRF-1 exerting negative feedback inhibition on STAT1 and downstream chemokine expression. The present study elucidates the transcriptional networks through which the IFN-γ/STAT1/IRF-1 axis controls β-cell function/differentiation, demise, and islet inflammation.
Figures




References
-
- Estruch F. (2000) FEMS Microbiol. Rev. 24, 469–486 - PubMed
-
- Eizirik D. L., Moore F., Flamez D., Ortis F. (2008) Biochem. Soc. Trans. 36, 321–327 - PubMed
-
- Gepts W. (1965) Diabetes 14, 619–633 - PubMed
-
- Cnop M., Welsh N., Jonas J. C., Jörns A., Lenzen S., Eizirik D. L. (2005) Diabetes 54, S97–107 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

