X-ray and biochemical analysis of N370S mutant human acid β-glucosidase
- PMID: 20980263
- PMCID: PMC3012987
- DOI: 10.1074/jbc.M110.150433
X-ray and biochemical analysis of N370S mutant human acid β-glucosidase
Abstract
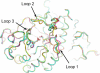
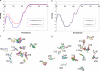
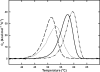
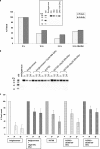
Gaucher disease is caused by mutations in the enzyme acid β-glucosidase (GCase), the most common of which is the substitution of serine for asparagine at residue 370 (N370S). To characterize the nature of this mutation, we expressed human N370S GCase in insect cells and compared the x-ray structure and biochemical properties of the purified protein with that of the recombinant human GCase (imiglucerase, Cerezyme®). The x-ray structure of N370S mutant acid β-glucosidase at acidic and neutral pH values indicates that the overall folding of the N370S mutant is identical to that of recombinant GCase. Subtle differences were observed in the conformation of a flexible loop at the active site and in the hydrogen bonding ability of aromatic residues on this loop with residue 370 and the catalytic residues Glu-235 and Glu-340. Circular dichroism spectroscopy showed a pH-dependent change in the environment of tryptophan residues in imiglucerase that is absent in N370S GCase. The mutant protein was catalytically deficient with reduced V(max) and increased K(m) values for the substrate p-nitrophenyl-β-D-glucopyranoside and reduced sensitivity to competitive inhibitors. N370S GCase was more stable to thermal denaturation and had an increased lysosomal half-life compared with imiglucerase following uptake into macrophages. The competitive inhibitor N-(n-nonyl)deoxynojirimycin increased lysosomal levels of both N370S and imiglucerase 2-3-fold by reducing lysosomal degradation. Overall, these data indicate that the N370S mutation results in a normally folded but less flexible protein with reduced catalytic activity compared with imiglucerase.
Figures
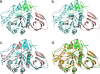
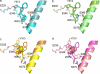
References
-
- Grabowski G. A., Petsko G. A., Kolodny E. H. (2006) in Metabolic and Molecular Bases of Inherited Disease (Scriver C. R., Beaudet A. L., Valle D., Sly W. S., Vogelstein B., Childs B., Kinzler K. W. eds) McGraw-Hill Inc., New York
-
- Beutler E., Gelbart T., Scott C. R. (2005) Blood Cells Mol. Dis. 35, 355–364 - PubMed
-
- Grace M. E., Graves P. N., Smith F. I., Grabowski G. A. (1990) J. Biol. Chem. 265, 6827–6835 - PubMed
-
- Grace M. E., Newman K. M., Scheinker V., Berg-Fussman A., Grabowski G. A. (1994) J. Biol. Chem. 269, 2283–2291 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

