DNA damage response
- PMID: 20980439
- PMCID: PMC3003462
- DOI: 10.1101/cshperspect.a000745
DNA damage response
Abstract
Structural changes to DNA severely affect its functions, such as replication and transcription, and play a major role in age-related diseases and cancer. A complicated and entangled network of DNA damage response (DDR) mechanisms, including multiple DNA repair pathways, damage tolerance processes, and cell-cycle checkpoints safeguard genomic integrity. Like transcription and replication, DDR is a chromatin-associated process that is generally tightly controlled in time and space. As DNA damage can occur at any time on any genomic location, a specialized spatio-temporal orchestration of this defense apparatus is required.
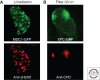
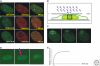
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
