Inositol hexakisphosphate kinase-2 acts as an effector of the vertebrate Hedgehog pathway
- PMID: 20980661
- PMCID: PMC2993352
- DOI: 10.1073/pnas.1007256107
Inositol hexakisphosphate kinase-2 acts as an effector of the vertebrate Hedgehog pathway
Abstract
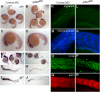
Inositol phosphate (IP) kinases constitute an emerging class of cellular kinases linked to multiple cellular activities. Here, we report a previously uncharacterized cellular function in Hedgehog (Hh) signaling for the IP kinase designated inositol hexakisphosphate kinase-2 (IP6K2) that produces diphosphoryl inositol phosphates (PP-IPs). In zebrafish embryos, IP6K2 activity was required for normal development of craniofacial structures, somites, and neural crest cells. ip6k2 depletion in both zebrafish and mammalian cells also inhibited Hh target gene expression. Inhibiting IP(6) kinase activity using N(2)-(m-(trifluoromethy)lbenzyl) N(6)-(p-nitrobenzyl)purine (TNP) resulted in altered Hh signal transduction. In zebrafish, restoring IP6K2 levels with exogenous ip6k2 mRNA reversed the effects of IP6K2 depletion. Furthermore, overexpression of ip6k2 in mammalian cells enhanced the Hh pathway response, suggesting IP6K2 is a positive regulator of Hh signaling. Perturbations from IP6K2 depletion or TNP were reversed by overexpressing smoM2, gli1, or ip6k2. Moreover, the inhibitory effect of cyclopamine was reversed by overexpressing ip6k2. This identified roles for the inositol kinase pathway in early vertebrate development and tissue morphogenesis, and in Hh signaling. We propose that IP6K2 activity is required at the level or downstream of Smoothened but upstream of the transcription activator Gli1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

