Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase
- PMID: 20980813
- PMCID: PMC3039730
- DOI: 10.4161/auto.7.1.13840
Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase
Abstract
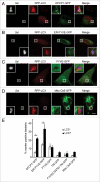
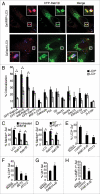
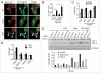
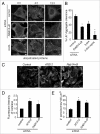
Autophagy mediates the degradation of cytoplasmic components in eukaryotic cells and plays a key role in immunity. The mechanism of autophagosome formation is not clear. Here we examined two potential membrane sources for antibacterial autophagy: the ER and mitochondria. DFCP1, a marker of specialized ER domains known as 'omegasomes,' associated with Salmonella-containing autophagosomes via its PtdIns(3)P and ER-binding domains, while a mitochondrial marker (cytochrome b5-GFP) did not. Rab1 also localized to autophagosomes, and its activity was required for autophagosome formation, clearance of protein aggregates and peroxisomes, and autophagy of Salmonella. Overexpression of Rab1 enhanced antibacterial autophagy. The role of Rab1 in antibacterial autophagy was independent of its role in ER-to-Golgi transport. Our data suggest that antibacterial autophagy occurs at omegasomes and reveal that the Rab1 GTPase plays a crucial role in mammalian autophagy.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
