A role for TREX components in the release of spliced mRNA from nuclear speckle domains
- PMID: 20981025
- PMCID: PMC3793428
- DOI: 10.1038/ncomms1103
A role for TREX components in the release of spliced mRNA from nuclear speckle domains
Abstract
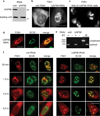
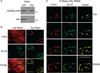
The TREX complex, which functions in mRNA export, is recruited to mRNA during splicing. Both the splicing machinery and the TREX complex are concentrated in 20-50 discrete foci known as nuclear speckle domains. In this study, we use a model system where DNA constructs are microinjected into HeLa cell nuclei, to follow the fates of the transcripts. We show that transcripts lacking functional splice sites, which are inefficiently exported, do not associate with nuclear speckle domains but are instead distributed throughout the nucleoplasm. In contrast, pre-mRNAs containing functional splice sites accumulate in nuclear speckles, and our data suggest that splicing occurs in these domains. When the TREX components UAP56 or Aly are knocked down, spliced mRNA, as well as total polyA+ RNA, accumulates in nuclear speckle domains. Together, our data raise the possibility that pre-mRNA undergoes splicing in nuclear speckle domains, before their release by TREX components for efficient export to the cytoplasm.
Figures
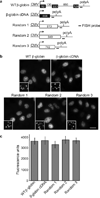
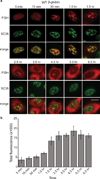
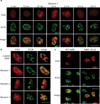
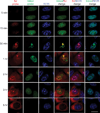


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

