Spatial organization of intestinal microbiota in the mouse ascending colon
- PMID: 20981114
- PMCID: PMC3105732
- DOI: 10.1038/ismej.2010.161
Spatial organization of intestinal microbiota in the mouse ascending colon
Abstract
Complex microbial populations are organized in relation to their environment. In the intestine, the inner lining (mucosa) is a potential focal point for such organization. The proximal murine colon contains mucosal folds that are known to be associated with morphologically distinct microbes. To identify these microbes, we used the technique of laser capture microdissection (LCM) to sample microbes associated with these folds (interfold region) and within the central lumen (digesta region). Using 16S rRNA gene tag pyrosequencing, we found that microbes in the interfold region were highly enriched for the phylum Firmicutes and, more specifically, for the families Lachnospiraceae and Ruminococcaceae. Other families such as Bacteroidaceae, Enterococcaceae and Lactobacillaceae were all enriched in the digesta region. This high-resolution system to capture and examine spatial organization of intestinal microbes should facilitate microbial analysis in other mouse models, furthering our understanding of host-microbial interactions.
Figures
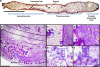
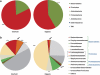

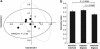

References
-
- Adekambi T, Shinnick TM, Raoult D, Drancourt M. Complete rpoB gene sequencing as a suitable supplement to DNA-DNA hybridization for bacterial species and genus delineation. Int J Syst Evol Microbiol. 2008;58:1807–1814. - PubMed
-
- Avaniss-Aghajani E, Jones K, Chapman D, Brunk C.1994A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences Biotechniques 17144–146.148–149. - PubMed
-
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. - PubMed
-
- Boye M, Aalbaek B, Agerholm JS. Fusobacterium necrophorum determined as abortifacient in sheep by laser capture microdissection and fluorescence in situ hybridization. Mol Cell Probes. 2006;20:330–336. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

