Germinal Cell Aplasia in Kif18a Mutant Male Mice Due to Impaired Chromosome Congression and Dysregulated BubR1 and CENP-E
- PMID: 20981276
- PMCID: PMC2963078
- DOI: 10.1177/1947601909358184
Germinal Cell Aplasia in Kif18a Mutant Male Mice Due to Impaired Chromosome Congression and Dysregulated BubR1 and CENP-E
Abstract
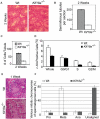
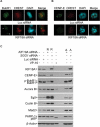
Chromosomal instability during cell division frequently causes cell death or malignant transformation. Orderly chromosome congression at the metaphase plate, a paramount process to vertebrate mitosis and meiosis, is controlled by a number of molecular regulators, including kinesins. Kinesin-8 (Kif18A) functions to control mitotic chromosome alignment at the mid-zone by negative regulation of kinetochore oscillation. Here the authors report that disrupting Kif18a function results in complete sterility in male but not in female mice. Histological examination reveals that Kif18a(-/-) testes exhibit severe developmental impairment of seminiferous tubules. Testis atrophy in Kif18a(-/-) mice is caused by perturbation of microtubule dynamics and spindle pole integrity, leading to chromosome congression defects during mitosis and meiosis. Depletion of KIF18A via RNAi causes mitotic arrest accompanied by unaligned chromosomes and increased microtubule nucleating centers in both GC-1 and HeLa cells. Prolonged depletion of KIF18A causes apoptosis due to perturbed microtubule dynamics. Further studies reveal that KIF18A silencing results in degradation of CENP-E and BubR1, which is accompanied by premature sister chromatid separation. KIF18A physically interacts with BubR1 and CENP-E, and this interaction is modulated during mitosis. Combined, the studies indicate that KIF18A is essential for normal chromosome congression during cell division and that the absence of KIF18A function causes severe defects in microtubule dynamics, spindle integrity, and checkpoint activation, leading to germinal cell aplasia in mice.
Keywords: BubR1; KIF18A; chromosome congression; knockout mice; testis development.
Conflict of interest statement
The authors declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
Figures





References
-
- Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol 2002;4(suppl):s41-9 - PubMed
-
- de Kretser DM. Male infertility. Lancet 1997;349:787-90 - PubMed
-
- Meschede D, Horst J. The molecular genetics of male infertility. Mol Hum Reprod 1997;3:419-30 - PubMed
-
- Kulczycki LL, Kostuch M, Bellanti JA. A clinical perspective of cystic fibrosis and new genetic findings: relationship of CFTR mutations to genotype-phenotype manifestations. Am J Med Genet 2003;116A:262-7 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
