Genome-wide analysis of phenylpropanoid defence pathways
- PMID: 21029326
- PMCID: PMC6640277
- DOI: 10.1111/j.1364-3703.2010.00648.x
Genome-wide analysis of phenylpropanoid defence pathways
Abstract
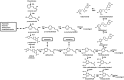
Phenylpropanoids can function as preformed and inducible antimicrobial compounds, as well as signal molecules, in plant-microbe interactions. Since we last reviewed the field 8 years ago, there has been a huge increase in our understanding of the genes of phenylpropanoid biosynthesis and their regulation, brought about largely by advances in genome technology, from whole-genome sequencing to massively parallel gene expression profiling. Here, we present an overview of the biosynthesis and roles of phenylpropanoids in plant defence, together with an analysis of confirmed and predicted phenylpropanoid pathway genes in the sequenced genomes of 11 plant species. Examples are provided of phylogenetic and expression clustering analyses, and the large body of underlying genomic data is provided through a website accessible from the article.
© 2010 The Authors.Molecular Plant Pathology © 2010 BSPP and Blackwell Publishing Ltd.
Figures






Similar articles
-
Phenylpropanoid Derivatives and Their Role in Plants' Health and as antimicrobials.Curr Microbiol. 2023 Oct 20;80(12):380. doi: 10.1007/s00284-023-03502-x. Curr Microbiol. 2023. PMID: 37864088 Review.
-
The phenylpropanoid pathway in Arabidopsis.Arabidopsis Book. 2011;9:e0152. doi: 10.1199/tab.0152. Epub 2011 Dec 6. Arabidopsis Book. 2011. PMID: 22303276 Free PMC article.
-
Lignin and lignans in plant defence: insight from expression profiling of cinnamyl alcohol dehydrogenase genes during development and following fungal infection in Populus.Plant Sci. 2014 Dec;229:111-121. doi: 10.1016/j.plantsci.2014.08.015. Epub 2014 Aug 29. Plant Sci. 2014. PMID: 25443838
-
Cytochromes P450 in phenylpropanoid metabolism.Drug Metabol Drug Interact. 1995;12(3-4):221-43. doi: 10.1515/dmdi.1995.12.3-4.221. Drug Metabol Drug Interact. 1995. PMID: 8820854 Review.
-
Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions.J Integr Plant Biol. 2021 Jan;63(1):180-209. doi: 10.1111/jipb.13054. J Integr Plant Biol. 2021. PMID: 33325112 Review.
Cited by
-
The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants.Mol Plant Pathol. 2019 Mar;20(3):309-322. doi: 10.1111/mpp.12755. Epub 2018 Nov 15. Mol Plant Pathol. 2019. PMID: 30267563 Free PMC article.
-
Understanding Host-Pathogen Interactions with Expression Profiling of NILs Carrying Rice-Blast Resistance Pi9 Gene.Front Plant Sci. 2017 Feb 23;8:93. doi: 10.3389/fpls.2017.00093. eCollection 2017. Front Plant Sci. 2017. PMID: 28280498 Free PMC article.
-
Microbial allies: Enhancing plant defense via phenylpropanoid pathway and lignification.Plant Physiol. 2025 Mar 1;197(3):kiaf059. doi: 10.1093/plphys/kiaf059. Plant Physiol. 2025. PMID: 40037578 Free PMC article. No abstract available.
-
Message hidden in α-helices-toward a better understanding of plant ABCG transporters' multispecificity.Plant Physiol. 2025 Apr 30;198(1):kiaf146. doi: 10.1093/plphys/kiaf146. Plant Physiol. 2025. PMID: 40220341 Free PMC article. No abstract available.
-
Commonly and Specifically Activated Defense Responses in Maize Disease Lesion Mimic Mutants Revealed by Integrated Transcriptomics and Metabolomics Analysis.Front Plant Sci. 2021 May 17;12:638792. doi: 10.3389/fpls.2021.638792. eCollection 2021. Front Plant Sci. 2021. PMID: 34079566 Free PMC article.
References
-
- Akashi, T. , Aoki, T. and Ayabe, S. (1998a) CYP81E1, a cytochrome P450 cDNA of licorice (Glycyrrhiza echinata L.), encodes isoflavone 2′‐hydroxylase. Biochem. Biophys. Res. Commun. 251, 67–70. - PubMed
-
- Akashi, T. , Aoki, T. and Ayabe, S. (1998b) Identification of a cytochrome P450 cDNA encoding (2S)‐flavanone 2‐hydroxylase of licorice (Glycyrrhiza echinata L.; Fabaceae) which represents licodione synthase and flavone synthase II. FEBS Lett. 431, 287–290. - PubMed
-
- Akashi, T. , Sawada, Y. , Shimada, N. , Sakurai, N. , Aoki, T. and Ayabe, S. (2003) cDNA cloning and biochemical characterization of S‐adenosyl‐L‐methionine:2,7,4′‐trihydroxyisoflavanone 4′‐O‐methyltransferase, a critical enzyme of the legume isoflavonoid phytoalexin pathway. Plant Cell Physiol. 44, 103–112. - PubMed
-
- Akashi, T. , VanEtten, H.D. , Sawada, Y. , Wasmann, C.C. , Uchiyama, H. and Ayabe, S. (2006) Catalytic specificity of pea O‐methyltransferases suggests gene duplication for (+)‐pisatin biosynthesis. Phytochemistry, 67, 2525–2530. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

