Transcriptional control in the prereplicative phase of T4 development
- PMID: 21029433
- PMCID: PMC2988021
- DOI: 10.1186/1743-422X-7-289
Transcriptional control in the prereplicative phase of T4 development
Abstract
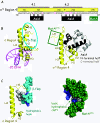
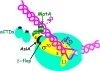
Control of transcription is crucial for correct gene expression and orderly development. For many years, bacteriophage T4 has provided a simple model system to investigate mechanisms that regulate this process. Development of T4 requires the transcription of early, middle and late RNAs. Because T4 does not encode its own RNA polymerase, it must redirect the polymerase of its host, E. coli, to the correct class of genes at the correct time. T4 accomplishes this through the action of phage-encoded factors. Here I review recent studies investigating the transcription of T4 prereplicative genes, which are expressed as early and middle transcripts. Early RNAs are generated immediately after infection from T4 promoters that contain excellent recognition sequences for host polymerase. Consequently, the early promoters compete extremely well with host promoters for the available polymerase. T4 early promoter activity is further enhanced by the action of the T4 Alt protein, a component of the phage head that is injected into E. coli along with the phage DNA. Alt modifies Arg265 on one of the two α subunits of RNA polymerase. Although work with host promoters predicts that this modification should decrease promoter activity, transcription from some T4 early promoters increases when RNA polymerase is modified by Alt. Transcription of T4 middle genes begins about 1 minute after infection and proceeds by two pathways: 1) extension of early transcripts into downstream middle genes and 2) activation of T4 middle promoters through a process called sigma appropriation. In this activation, the T4 co-activator AsiA binds to Region 4 of σ⁷⁰, the specificity subunit of RNA polymerase. This binding dramatically remodels this portion of σ⁷⁰, which then allows the T4 activator MotA to also interact with σ⁷⁰. In addition, AsiA restructuring of σ⁷⁰ prevents Region 4 from forming its normal contacts with the -35 region of promoter DNA, which in turn allows MotA to interact with its DNA binding site, a MotA box, centered at the -30 region of middle promoter DNA. T4 sigma appropriation reveals how a specific domain within RNA polymerase can be remolded and then exploited to alter promoter specificity.
Figures
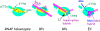

References
-
- Stitt B, Hinton DM. In: Molecular biology of bacteriophage T4. Karam JD, Drake J, Kreuzer KN, Mosig G, Hall D, Eiserling F, Black L, Spicer E, Kutter E, Carlson K, Miller ES, editor. Washington, D.C.: American Society for Microbiology; 1994. Regulation of middle-mode transcription; pp. 142–160.
-
- Hinton DM, Pande S, Wais N, Johnson XB, Vuthoori M, Makela A, Hook-Barnard I. Transcriptional takeover by sigma appropriation: remodelling of the sigma70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA. Microbiology. 2005;151:1729–1740. doi: 10.1099/mic.0.27972-0. - DOI - PubMed
-
- Brody E, Rabussay D, Hall D. In: Bacteriophage T4. Mathews CK, Kutter EM, Mosig G, Berget PB, editor. Washington, D. C.: American Society for Microbiology; 1983. Regulation of transcription of prereplicative genes; pp. 174–183.
-
- Wilkens K, Ruger W. In: Molecular Biology of Bacteriophage T4. Karam JD, Drake JW, Kreuzer KN, Mosig G, Hall DH, Eiserling FA, Black LW, Spicer EK, Kutter E, Carlson K, Miller ES, editor. Washington, D. C.: American Society for Microbiology; 1994. Transcription from early promoters; pp. 132–141.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

