Mobile DNA elements in T4 and related phages
- PMID: 21029434
- PMCID: PMC2988022
- DOI: 10.1186/1743-422X-7-290
Mobile DNA elements in T4 and related phages
Abstract
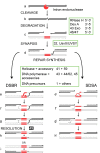
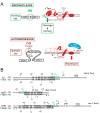
Mobile genetic elements are common inhabitants of virtually every genome where they can exert profound influences on genome structure and function in addition to promoting their own spread within and between genomes. Phage T4 and related phage have long served as a model system for understanding the molecular mechanisms by which a certain class of mobile DNA, homing endonucleases, promote their spread. Homing endonucleases are site-specific DNA endonucleases that initiate mobility by introducing double-strand breaks at defined positions in genomes lacking the endonuclease gene, stimulating repair and recombination pathways that mobilize the endonuclease coding region. In phage T4, homing endonucleases were first discovered as encoded within the self-splicing td, nrdB and nrdD introns of T4. Genomic data has revealed that homing endonucleases are extremely widespread in T-even-like phage, as evidenced by the astounding fact that ~11% of the T4 genome encodes homing endonuclease genes, with most of them located outside of self-splicing introns. Detailed studies of the mobile td intron and its encoded endonuclease, I-TevI, have laid the foundation for genetic, biochemical and structural aspects that regulate the mobility process, and more recently have provided insights into regulation of homing endonuclease function. Here, we summarize the current state of knowledge regarding T4-encoded homing endonucleases, with particular emphasis on the td/I-TevI model system. We also discuss recent progress in the biology of free-standing endonucleases, and present areas of future research for this fascinating class of mobile genetic elements.
Figures


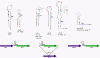
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

