MicroR159 regulation of most conserved targets in Arabidopsis has negligible phenotypic effects
- PMID: 21029441
- PMCID: PMC2988730
- DOI: 10.1186/1758-907X-1-18
MicroR159 regulation of most conserved targets in Arabidopsis has negligible phenotypic effects
Abstract
Background: A current challenge of microRNA (miRNA) research is the identification of biologically relevant miRNA:target gene relationships. In plants, high miRNA:target gene complementarity has enabled accurate target predictions, and slicing of target mRNAs has facilitated target validation through rapid amplification of 5' cDNA ends (5'-RACE) analysis. Together, these approaches have identified more than 20 targets potentially regulated by the deeply conserved miR159 family in Arabidopsis, including eight MYB genes with highly conserved miR159 target sites. However, genetic analysis has revealed the functional specificity of the major family members, miR159a and miR159b is limited to only two targets, MYB33 and MYB65. Here, we examine the functional role of miR159 regulation for the other potential MYB target genes.
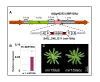
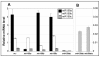
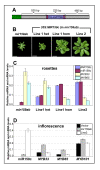
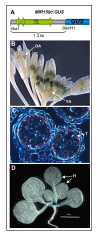
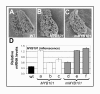
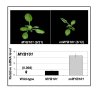
Results: For these target genes, functional analysis failed to identify miR159 regulation that resulted in any major phenotypic impact, either at the morphological or molecular level. This appears to be mainly due to the quiescent nature of the remaining family member, MIR159c. Although its expression overlaps in a temporal and spatial cell-specific manner with a subset of these targets in anthers, the abundance of miR159c is extremely low and concomitantly a mir159c mutant displays no anther defects. Examination of potential miR159c targets with conserved miR159 binding sites found neither their spatial or temporal expression domains appeared miR159 regulated, despite the detection of miR159-guided cleavage products by 5'-RACE. Moreover, expression of a miR159-resistant target (mMYB101) resulted predominantly in plants that are indistinguishable from wild type. Plants that displayed altered morphological phenotypes were found to be ectopically expressing the mMYB101 transgene, and hence were misrepresentative of the in vivo functional role of miR159.
Conclusions: This study presents a novel explanation for a paradox common to plant and animal miRNA systems, where among many potential miRNA-target relationships usually only a few appear physiologically relevant. The identification of a quiescent miR159c:target gene regulatory module in anthers provides a likely rationale for the presence of conserved miR159 binding sites in many targets for which miR159 regulation has no obvious functional role. Remnants from the demise of such modules may lead to an overestimation of miRNA regulatory complexity when investigated using bioinformatic, 5'-RACE or transgenic approaches.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
