Genomic analysis of microRNA time-course expression in liver of mice treated with genotoxic carcinogen N-ethyl-N-nitrosourea
- PMID: 21029445
- PMCID: PMC3091750
- DOI: 10.1186/1471-2164-11-609
Genomic analysis of microRNA time-course expression in liver of mice treated with genotoxic carcinogen N-ethyl-N-nitrosourea
Abstract
Background: Dysregulated expression of microRNAs (miRNAs) has been previously observed in human cancer tissues and shown promise in defining tumor status. However, there is little information as to if or when expression changes of miRNAs occur in normal tissues after carcinogen exposure.
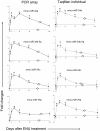
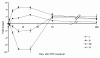
Results: To explore the possible time-course changes of miRNA expression induced by a carcinogen, we treated mice with one dose of 120 mg/kg N-ethyl-N-nitrosourea (ENU), a model genotoxic carcinogen, and vehicle control. The miRNA expression profiles were assessed in the mouse livers in a time-course design. miRNAs were isolated from the livers at days 1, 3, 7, 15, 30 and 120 after the treatment and their expression was determined using a miRNA PCR Array. Principal component analysis of the miRNA expression profiles showed that miRNA expression at post-treatment days (PTDs) 7 and 15 were different from those at the other time points and the control. The number of differentially expressed miRNAs (DEMs) changed over time (3, 5, 14, 32, 5 and 5 at PTDs 1, 3, 7, 15, 30 and 120, respectively). The magnitude of the expression change varied with time with the highest changes at PTDs 7 or 15 for most of the DEMs. In silico functional analysis of the DEMs at PTDs 7 and 15 indicated that the major functions of these ENU-induced DEMs were associated with DNA damage, DNA repair, apoptosis and other processes related to carcinogenesis.
Conclusion: Our results showed that many miRNAs changed their expression to respond the exposure of the genotoxic carcinogen ENU and the number and magnitude of the changes were highest at PTDs 7 to 15. Thus, one to two weeks after the exposure is the best time for miRNA expression sampling.
Figures



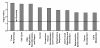
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

