Nucleosome-interacting proteins regulated by DNA and histone methylation
- PMID: 21029866
- PMCID: PMC3640253
- DOI: 10.1016/j.cell.2010.10.012
Nucleosome-interacting proteins regulated by DNA and histone methylation
Abstract
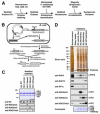
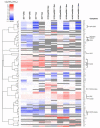
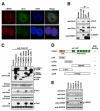
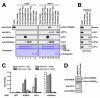
Modifications on histones or on DNA recruit proteins that regulate chromatin function. Here, we use nucleosomes methylated on DNA and on histone H3 in an affinity assay, in conjunction with a SILAC-based proteomic analysis, to identify "crosstalk" between these two distinct classes of modification. Our analysis reveals proteins whose binding to nucleosomes is regulated by methylation of CpGs, H3K4, H3K9, and H3K27 or a combination thereof. We identify the origin recognition complex (ORC), including LRWD1 as a subunit, to be a methylation-sensitive nucleosome interactor that is recruited cooperatively by DNA and histone methylation. Other interactors, such as the lysine demethylase Fbxl11/KDM2A, recognize nucleosomes methylated on histones, but their recruitment is disrupted by DNA methylation. These data establish SILAC nucleosome affinity purifications (SNAP) as a tool for studying the dynamics between different chromatin modifications and provide a modification binding "profile" for proteins regulated by DNA and histone methylation.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

References
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
-
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

