HDAC3 is negatively regulated by the nuclear protein DBC1
- PMID: 21030595
- PMCID: PMC3003384
- DOI: 10.1074/jbc.M110.153270
HDAC3 is negatively regulated by the nuclear protein DBC1
Abstract
HDAC3 is a member of the class I histone deacetylase family that regulates gene expression by deacetylation of histones and non-histone proteins. HDAC3 activity has been shown to be modulated by interaction with the co-repressors NCoR and SMRT. Here, we present evidence that the nuclear protein DBC1 is an endogenous inhibitor of HDAC3. DBC1 has been previously identified as a regulator of some nuclear receptors, the methyltransferase SUV39H1, and the NAD-dependent deacetylase SIRT1. Furthermore, DBC1 has been shown to influence transcription regulation and apoptosis, and it may also act as a tumor suppressor. We found that DBC1 interacts and specifically inhibits the deacetylase HDAC3. This interaction depends on the N terminus of DBC1 and the C terminus of HDAC3. Expression of DBC1 not only inhibited HDAC3 activity but also altered its subcellular distribution. In addition, knockdown of endogenous DBC1 in cells and knock-out in mouse tissues increased HDAC3 deacetylase activity. Together, these results identify DBC1 as a new regulator of HDAC3 and demonstrate that DBC1 is a negative regulator of two key distinct deacetylases, SIRT1 and HDAC3. These findings may lead to a better understanding of the biological roles of DBC1 and HDAC3 in metabolic diseases and cancer.
Figures


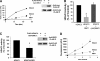
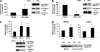


Similar articles
-
Inhibition of SUV39H1 methyltransferase activity by DBC1.J Biol Chem. 2009 Apr 17;284(16):10361-6. doi: 10.1074/jbc.M900956200. Epub 2009 Feb 13. J Biol Chem. 2009. PMID: 19218236 Free PMC article.
-
Role of deleted in breast cancer 1 (DBC1) protein in SIRT1 deacetylase activation induced by protein kinase A and AMP-activated protein kinase.J Biol Chem. 2012 Jul 6;287(28):23489-501. doi: 10.1074/jbc.M112.365874. Epub 2012 May 2. J Biol Chem. 2012. PMID: 22553202 Free PMC article.
-
DBC1/CCAR2 and CCAR1 Are Largely Disordered Proteins that Have Evolved from One Common Ancestor.Biomed Res Int. 2014;2014:418458. doi: 10.1155/2014/418458. Epub 2014 Dec 11. Biomed Res Int. 2014. PMID: 25610865 Free PMC article.
-
Cell cycle and apoptosis regulator 2 at the interface between DNA damage response and cell physiology.Mutat Res Rev Mutat Res. 2018 Apr-Jun;776:1-9. doi: 10.1016/j.mrrev.2018.03.004. Epub 2018 Mar 19. Mutat Res Rev Mutat Res. 2018. PMID: 29807573 Review.
-
Integrative regulation of physiology by histone deacetylase 3.Nat Rev Mol Cell Biol. 2019 Feb;20(2):102-115. doi: 10.1038/s41580-018-0076-0. Nat Rev Mol Cell Biol. 2019. PMID: 30390028 Free PMC article. Review.
Cited by
-
Breast cancer metastasis suppressor 1 modulates SIRT1-dependent p53 deacetylation through interacting with DBC1.Am J Cancer Res. 2016 Jun 1;6(6):1441-9. eCollection 2016. Am J Cancer Res. 2016. PMID: 27429856 Free PMC article.
-
Hypoxia-induced proteasomal degradation of DBC1 by SIAH2 in breast cancer progression.Elife. 2022 Aug 1;11:e81247. doi: 10.7554/eLife.81247. Elife. 2022. PMID: 35913115 Free PMC article.
-
Emerging Roles of p53 Related lncRNAs in Cancer Progression: A Systematic Review.Int J Biol Sci. 2019 May 12;15(6):1287-1298. doi: 10.7150/ijbs.33218. eCollection 2019. Int J Biol Sci. 2019. PMID: 31223287 Free PMC article.
-
Deleted in breast cancer 1 limits adipose tissue fat accumulation and plays a key role in the development of metabolic syndrome phenotype.Diabetes. 2015 Jan;64(1):12-22. doi: 10.2337/db14-0192. Epub 2014 Jul 22. Diabetes. 2015. PMID: 25053585 Free PMC article.
-
DBC1 promotes castration-resistant prostate cancer by positively regulating DNA binding and stability of AR-V7.Oncogene. 2018 Mar;37(10):1326-1339. doi: 10.1038/s41388-017-0047-5. Epub 2017 Dec 18. Oncogene. 2018. PMID: 29249800
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

