Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit
- PMID: 21030658
- PMCID: PMC2990404
- DOI: 10.1126/science.1193220
Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit
Abstract
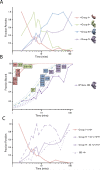
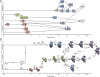
Ribosomes are self-assembling macromolecular machines that translate DNA into proteins, and an understanding of ribosome biogenesis is central to cellular physiology. Previous studies on the Escherichia coli 30S subunit suggest that ribosome assembly occurs via multiple parallel pathways rather than through a single rate-limiting step, but little mechanistic information is known about this process. Discovery single-particle profiling (DSP), an application of time-resolved electron microscopy, was used to obtain more than 1 million snapshots of assembling 30S subunits, identify and visualize the structures of 14 assembly intermediates, and monitor the population flux of these intermediates over time. DSP results were integrated with mass spectrometry data to construct the first ribosome-assembly mechanism that incorporates binding dependencies, rate constants, and structural characterization of populated intermediates.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

