RISC RNA sequencing for context-specific identification of in vivo microRNA targets
- PMID: 21030712
- PMCID: PMC3017647
- DOI: 10.1161/CIRCRESAHA.110.233528
RISC RNA sequencing for context-specific identification of in vivo microRNA targets
Abstract
Rationale: MicroRNAs (miRs) are expanding our understanding of cardiac disease and have the potential to transform cardiovascular therapeutics. One miR can target hundreds of individual mRNAs, but existing methodologies are not sufficient to accurately and comprehensively identify these mRNA targets in vivo.
Objective: To develop methods permitting identification of in vivo miR targets in an unbiased manner, using massively parallel sequencing of mouse cardiac transcriptomes in combination with sequencing of mRNA associated with mouse cardiac RNA-induced silencing complexes (RISCs).
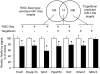
Methods and results: We optimized techniques for expression profiling small amounts of RNA without introducing amplification bias and applied this to anti-Argonaute 2 immunoprecipitated RISCs (RISC-Seq) from mouse hearts. By comparing RNA-sequencing results of cardiac RISC and transcriptome from the same individual hearts, we defined 1645 mRNAs consistently targeted to mouse cardiac RISCs. We used this approach in hearts overexpressing miRs from Myh6 promoter-driven precursors (programmed RISC-Seq) to identify 209 in vivo targets of miR-133a and 81 in vivo targets of miR-499. Consistent with the fact that miR-133a and miR-499 have widely differing "seed" sequences and belong to different miR families, only 6 targets were common to miR-133a- and miR-499-programmed hearts.
Conclusions: RISC-sequencing is a highly sensitive method for general RISC profiling and individual miR target identification in biological context and is applicable to any tissue and any disease state.
Figures


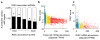


Comment in
-
"RISCing" the heart: In vivo identification of cardiac microRNA targets by RISCome.Circ Res. 2011 Jan 7;108(1):3-5. doi: 10.1161/CIRCRESAHA.110.238584. Circ Res. 2011. PMID: 21212388 Free PMC article. No abstract available.
Similar articles
-
"RISCing" the heart: In vivo identification of cardiac microRNA targets by RISCome.Circ Res. 2011 Jan 7;108(1):3-5. doi: 10.1161/CIRCRESAHA.110.238584. Circ Res. 2011. PMID: 21212388 Free PMC article. No abstract available.
-
Cardiac Disease Status Dictates Functional mRNA Targeting Profiles of Individual MicroRNAs.Circ Cardiovasc Genet. 2015 Dec;8(6):774-84. doi: 10.1161/CIRCGENETICS.115.001237. Epub 2015 Nov 9. Circ Cardiovasc Genet. 2015. PMID: 26553694 Free PMC article.
-
Exploring miRNA-mRNA regulatory network in cardiac pathology in Na+/H+ exchanger isoform 1 transgenic mice.Physiol Genomics. 2018 Oct 1;50(10):846-861. doi: 10.1152/physiolgenomics.00048.2018. Epub 2018 Jul 20. Physiol Genomics. 2018. PMID: 30029588 Free PMC article.
-
siRNA and miRNA: an insight into RISCs.Trends Biochem Sci. 2005 Feb;30(2):106-14. doi: 10.1016/j.tibs.2004.12.007. Trends Biochem Sci. 2005. PMID: 15691656 Review.
-
Life of RISC: Formation, action, and degradation of RNA-induced silencing complex.Mol Cell. 2022 Jan 6;82(1):30-43. doi: 10.1016/j.molcel.2021.11.026. Epub 2021 Dec 22. Mol Cell. 2022. PMID: 34942118 Review.
Cited by
-
MicroRNAs and the butterfly effect.Cell Cycle. 2013 Mar 1;12(5):707-8. doi: 10.4161/cc.23858. Epub 2013 Feb 19. Cell Cycle. 2013. PMID: 23422861 Free PMC article. No abstract available.
-
"RISCing" the heart: In vivo identification of cardiac microRNA targets by RISCome.Circ Res. 2011 Jan 7;108(1):3-5. doi: 10.1161/CIRCRESAHA.110.238584. Circ Res. 2011. PMID: 21212388 Free PMC article. No abstract available.
-
Decoding the complex genetic causes of heart diseases using systems biology.Biophys Rev. 2015 Mar;7(1):141-159. doi: 10.1007/s12551-014-0145-3. Epub 2014 Dec 10. Biophys Rev. 2015. PMID: 28509974 Free PMC article.
-
Technologies to Study Genetics and Molecular Pathways.Adv Exp Med Biol. 2024;1441:435-458. doi: 10.1007/978-3-031-44087-8_22. Adv Exp Med Biol. 2024. PMID: 38884724 Review.
-
Induced pluripotent stem cells in cardiovascular drug discovery.Circ Res. 2013 Feb 1;112(3):534-48. doi: 10.1161/CIRCRESAHA.111.250266. Circ Res. 2013. PMID: 23371902 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

