Cryoprotective mechanism of a small intrinsically disordered dehydrin protein
- PMID: 21031484
- PMCID: PMC3047060
- DOI: 10.1002/pro.534
Cryoprotective mechanism of a small intrinsically disordered dehydrin protein
Abstract
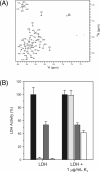
Dehydration proteins (Dehydrins) are expressed during dehydration stress in plants and are thought to protect plant proteins and membranes from the loss of water during drought and at cold temperatures. Several different dehydrins have been shown to protect lactate dehydrogenase (LDH) from damage from being frozen and thawed. We show here that a 48 residue K₂ dehydrin from Vitis riparia protects LDH more effectively than bovine serum albumin, a protein with known cryoprotective function. Light scattering and 8-anilino-1-naphthalene sulfonate fluorescence experiments show that dehydrins prevent aggregation and unfolding of the enzyme. The cryoprotective effects of LDH are reduced by the addition of salt, suggesting that the positively charged K-segments are attracted to a negatively charged surface but this does not result in binding. Overall K₂ is an intrinsically disordered protein; nuclear magnetic resonance relaxation experiments indicate that the two-terminal, Lys-rich K-segments show a weak propensity for α-helicity and are flexible, and that the central, polar rich phi-segment has no secondary structure preference and is highly flexible. We propose that the phi-segments in dehydrins are important for maintaining the disordered structure so that the protein can act as a molecular shield to prevent partially denatured proteins from interacting with one another, whereas the K-segments may help to localize the dehydrin near the enzyme surface.
Figures

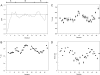

 , where x is the concentration of the additive.
, where x is the concentration of the additive.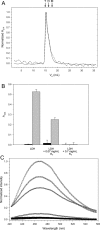
References
-
- Close TJ. Dehydrins: a commonality in the response of plants to dehydration and low temperature. Physiol Plant. 1997;100:291–296.
-
- Kosova K, Vitamvas P, Prasil IT. The role of dehydrins in plant response to cold. Biol Plant. 2007;51:601–617.
-
- Allagulova CR, Gimalov FR, Shakirova FM, Vakhitov VA. The plant dehydrins: structure and putative functions. Biochemistry. 2003;68:945–951. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

