Exploiting physico-chemical properties in string kernels
- PMID: 21034432
- PMCID: PMC2966294
- DOI: 10.1186/1471-2105-11-S8-S7
Exploiting physico-chemical properties in string kernels
Abstract
Background: String kernels are commonly used for the classification of biological sequences, nucleotide as well as amino acid sequences. Although string kernels are already very powerful, when it comes to amino acids they have a major short coming. They ignore an important piece of information when comparing amino acids: the physico-chemical properties such as size, hydrophobicity, or charge. This information is very valuable, especially when training data is less abundant. There have been only very few approaches so far that aim at combining these two ideas.
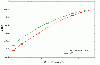
Results: We propose new string kernels that combine the benefits of physico-chemical descriptors for amino acids with the ones of string kernels. The benefits of the proposed kernels are assessed on two problems: MHC-peptide binding classification using position specific kernels and protein classification based on the substring spectrum of the sequences. Our experiments demonstrate that the incorporation of amino acid properties in string kernels yields improved performances compared to standard string kernels and to previously proposed non-substring kernels.
Conclusions: In summary, the proposed modifications, in particular the combination with the RBF substring kernel, consistently yield improvements without affecting the computational complexity. The proposed kernels therefore appear to be the kernels of choice for any protein sequence-based inference.
Availability: Data sets, code and additional information are available from http://www.fml.tuebingen.mpg.de/raetsch/suppl/aask. Implementations of the developed kernels are available as part of the Shogun toolbox.
Figures
Similar articles
-
A weighted string kernel for protein fold recognition.BMC Bioinformatics. 2017 Aug 25;18(1):378. doi: 10.1186/s12859-017-1795-5. BMC Bioinformatics. 2017. PMID: 28841820 Free PMC article.
-
Semi-supervised protein classification using cluster kernels.Bioinformatics. 2005 Aug 1;21(15):3241-7. doi: 10.1093/bioinformatics/bti497. Epub 2005 May 19. Bioinformatics. 2005. PMID: 15905279
-
Protein homology detection using string alignment kernels.Bioinformatics. 2004 Jul 22;20(11):1682-9. doi: 10.1093/bioinformatics/bth141. Epub 2004 Feb 26. Bioinformatics. 2004. PMID: 14988126
-
Learned random-walk kernels and empirical-map kernels for protein sequence classification.J Comput Biol. 2009 Mar;16(3):457-74. doi: 10.1089/cmb.2008.0031. J Comput Biol. 2009. PMID: 19254184
-
Sequence-based protein superfamily classification using computational intelligence techniques: a review.Int J Data Min Bioinform. 2015;11(4):424-57. doi: 10.1504/ijdmb.2015.067957. Int J Data Min Bioinform. 2015. PMID: 26336668 Review.
Cited by
-
Machine learning assisted design of highly active peptides for drug discovery.PLoS Comput Biol. 2015 Apr 7;11(4):e1004074. doi: 10.1371/journal.pcbi.1004074. eCollection 2015 Apr. PLoS Comput Biol. 2015. PMID: 25849257 Free PMC article.
-
On learning functions over biological sequence space: relating Gaussian process priors, regularization, and gauge fixing.bioRxiv [Preprint]. 2025 Aug 2:2025.04.26.650699. doi: 10.1101/2025.04.26.650699. bioRxiv. 2025. PMID: 40672195 Free PMC article. Preprint.
-
Encodings and models for antimicrobial peptide classification for multi-resistant pathogens.BioData Min. 2019 Mar 4;12:7. doi: 10.1186/s13040-019-0196-x. eCollection 2019. BioData Min. 2019. PMID: 30867681 Free PMC article. Review.
-
On learning functions over biological sequence space: relating Gaussian process priors, regularization, and gauge fixing.ArXiv [Preprint]. 2025 Aug 1:arXiv:2504.19034v3. ArXiv. 2025. PMID: 40671954 Free PMC article. Preprint.
-
A weighted string kernel for protein fold recognition.BMC Bioinformatics. 2017 Aug 25;18(1):378. doi: 10.1186/s12859-017-1795-5. BMC Bioinformatics. 2017. PMID: 28841820 Free PMC article.
References
-
- Kuang R, Ie E, Wang K, Wang K, Siddiqi M, Freund Y, Leslie C. Profile-based string kernels for remote homology detection and motif extraction. Proceedings IEEE Computational Systems Bioinformatics Conference. 2004. - PubMed
-
- Schweikert G, Zien A, Zeller G, Behr J, Dieterich C, Ong CS, Philips P, De Bona F, Hartmann L, Bohlen A, Krüger N, Sonnenburg S, Ratsch G. mGene: accurate SVM-based gene finding with an application to nematode genomes. Genome Res. 2009;19(11):2133–43. doi: 10.1101/gr.090597.108. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

