Smoking-mediated up-regulation of GAD67 expression in the human airway epithelium
- PMID: 21034448
- PMCID: PMC2988726
- DOI: 10.1186/1465-9921-11-150
Smoking-mediated up-regulation of GAD67 expression in the human airway epithelium
Abstract
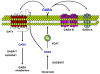
Background: The production of gamma-amino butyric acid (GABA) is dependent on glutamate decarboxylases (GAD65 and GAD67), the enzymes that catalyze the decarboxylation of glutamate to GABA. Based on studies suggesting a role of the airway epithelial GABAergic system in asthma-related mucus overproduction, we hypothesized that cigarette smoking, another disorder associated with increased mucus production, may modulate GABAergic system-related gene expression levels in the airway epithelium.
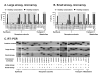
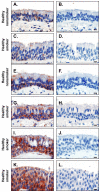
Methods: We assessed expression of the GABAergic system in human airway epithelium obtained using bronchoscopy to sample the epithelium and microarrays to evaluate gene expression. RT-PCR was used to confirm gene expression of GABAergic system gene in large and small airway epithelium from heathy nonsmokers and healthy smokers. The differences in the GABAergic system gene was further confirmed by TaqMan, immunohistochemistry and Western analysis.
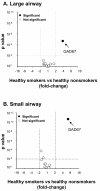
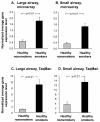
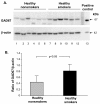
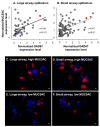
Results: The data demonstrate there is a complete GABAergic system expressed in the large and small human airway epithelium, including glutamate decarboxylase, GABA receptors, transporters and catabolism enzymes. Interestingly, of the entire GABAergic system, smoking modified only the expression of GAD67, with marked up-regulation of GAD67 gene expression in both large (4.1-fold increase, p < 0.01) and small airway epithelium of healthy smokers (6.3-fold increase, p < 0.01). At the protein level, Western analysis confirmed the increased expression of GAD67 in airway epithelium of healthy smokers compared to healthy nonsmokers (p < 0.05). There was a significant positive correlation between GAD67 and MUC5AC gene expression in both large and small airway epithelium (p < 0.01), implying a link between GAD67 and mucin overproduction in association with smoking.
Conclusions: In the context that GAD67 is the rate limiting enzyme in GABA synthesis, the correlation of GAD67 gene expression with MUC5AC expressions suggests that the up-regulation of airway epithelium expression of GAD67 may contribute to the increase in mucus production observed in association with cigarette smoking.
Trial registration: NCT00224198; NCT00224185.
Figures
References
-
- Watanabe M, Maemura K, Kanbara K, Tamayama T, Hayasaki H. GABA and GABA receptors in the central nervous system and other organs. Int Rev Cytol. 2002;213:1–47. full_text. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

