CoNSEnsX: an ensemble view of protein structures and NMR-derived experimental data
- PMID: 21034466
- PMCID: PMC2987814
- DOI: 10.1186/1472-6807-10-39
CoNSEnsX: an ensemble view of protein structures and NMR-derived experimental data
Abstract
Background: In conjunction with the recognition of the functional role of internal dynamics of proteins at various timescales, there is an emerging use of dynamic structural ensembles instead of individual conformers. These ensembles are usually substantially more diverse than conventional NMR ensembles and eliminate the expectation that a single conformer should fulfill all NMR parameters originating from 10(16) - 10(17) molecules in the sample tube. Thus, the accuracy of dynamic conformational ensembles should be evaluated differently to that of single conformers.
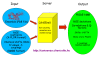
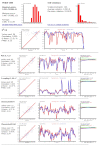
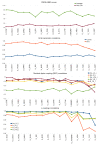
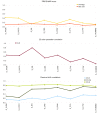
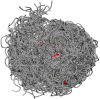
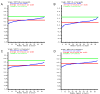
Results: We constructed the web application CoNSEnsX (Consistency of NMR-derived Structural Ensembles with eXperimental data) allowing fast, simple and convenient assessment of the correspondence of the ensemble as a whole with diverse independent NMR parameters available. We have chosen different ensembles of three proteins, human ubiquitin, a small protease inhibitor and a disordered subunit of cGMP phosphodiesterase 5/6 for detailed evaluation and demonstration of the capabilities of the CoNSEnsX approach.
Conclusions: Our results present a new conceptual method for the evaluation of dynamic conformational ensembles resulting from NMR structure determination. The designed CoNSEnsX approach gives a complete evaluation of these ensembles and is freely available as a web service at http://consensx.chem.elte.hu.
Figures
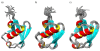
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

