Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species
- PMID: 21034474
- PMCID: PMC3218663
- DOI: 10.1186/gb-2010-11-10-r107
Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species
Erratum in
- Genome Biol. 2011;12(10):140
Abstract
Background: Streptococcus pneumoniae is one of the most important causes of microbial diseases in humans. The genomes of 44 diverse strains of S. pneumoniae were analyzed and compared with strains of non-pathogenic streptococci of the Mitis group.
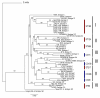
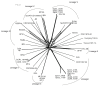
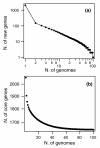
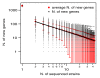
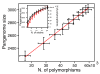
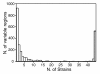
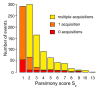
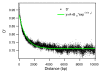
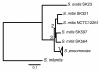
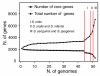
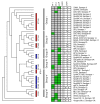
Results: Despite evidence of extensive recombination, the S. pneumoniae phylogenetic tree revealed six major lineages. With the exception of serotype 1, the tree correlated poorly with capsular serotype, geographical site of isolation and disease outcome. The distribution of dispensable genes--genes present in more than one strain but not in all strains--was consistent with phylogeny, although horizontal gene transfer events attenuated this correlation in the case of ancient lineages. Homologous recombination, involving short stretches of DNA, was the dominant evolutionary process of the core genome of S. pneumoniae. Genetic exchange occurred both within and across the borders of the species, and S. mitis was the main reservoir of genetic diversity of S. pneumoniae. The pan-genome size of S. pneumoniae increased logarithmically with the number of strains and linearly with the number of polymorphic sites of the sampled genomes, suggesting that acquired genes accumulate proportionately to the age of clones. Most genes associated with pathogenicity were shared by all S. pneumoniae strains, but were also present in S. mitis, S. oralis and S. infantis, indicating that these genes are not sufficient to determine virulence.
Conclusions: Genetic exchange with related species sharing the same ecological niche is the main mechanism of evolution of S. pneumoniae. The open pan-genome guarantees the species a quick and economical response to diverse environments.
Figures
References
-
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarity Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N. et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. - DOI - PMC - PubMed
-
- Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, Powell E, Keefe R, Ehrlich NE, Shen K, Hayes J, Barbadora K, Klimke W, Dernovoy D, Tatusova T, Parkhill J, Bentley SD, Post JC, Ehrlich GD, Hu FZ. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J Bacteriol. 2007;189:8186–8195. doi: 10.1128/JB.00690-07. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

