Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses
- PMID: 21035178
- PMCID: PMC3005334
- DOI: 10.1016/j.cell.2010.09.048
Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses
Abstract
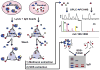
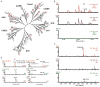
Natural small compounds comprise most cellular molecules and bind proteins as substrates, products, cofactors, and ligands. However, a large-scale investigation of in vivo protein-small metabolite interactions has not been performed. We developed a mass spectrometry assay for the large-scale identification of in vivo protein-hydrophobic small metabolite interactions in yeast and analyzed compounds that bind ergosterol biosynthetic proteins and protein kinases. Many of these proteins bind small metabolites; a few interactions were previously known, but the vast majority are new. Importantly, many key regulatory proteins such as protein kinases bind metabolites. Ergosterol was found to bind many proteins and may function as a general regulator. It is required for the activity of Ypk1, a mammalian AKT/SGK kinase homolog. Our study defines potential key regulatory steps in lipid biosynthetic pathways and suggests that small metabolites may play a more general role as regulators of protein activity and function than previously appreciated.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures



References
-
- Alberts B. Molecular biology of the cell. 4. New York: Garland Science; 2002.
-
- Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, Oliver SG, Kell DB. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol. 2003;21:692–696. - PubMed
-
- Babu P, Deschenes RJ, Robinson LC. Akr1p-dependent palmitoylation of Yck2p yeast casein kinase 1 is necessary and sufficient for plasma membrane targeting. J Biol Chem. 2004;279:27138–27147. - PubMed
-
- Beloqui A, Guazzaroni ME, Pazos F, Vieites JM, Godoy M, Golyshina OV, Chernikova TN, Waliczek A, Silva-Rocha R, Al-ramahi Y, et al. Reactome Array: Forging a Link Between Metabolome and Genome. Science. 2009;326:252–257. - PubMed
-
- Brazil DP, Hemmings BA. Ten years of protein kinase B signalling: a hard Akt to follow. Trends Biochem Sci. 2001;26:657–664. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

