Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks
- PMID: 21035407
- PMCID: PMC3008338
- DOI: 10.1016/j.dnarep.2010.10.001
Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks
Abstract
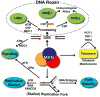
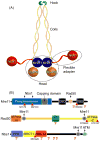
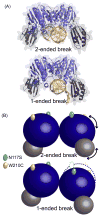
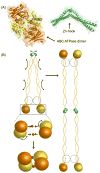
Repair and integrity of DNA ends at breaks, replication forks and telomeres are essential for life; yet, paradoxically, these responses are, in many cases, controlled by a single protein complex, Mre11-Rad50-Nbs1 (MRN). The MRN complex consists of dimers of each subunit and this heterohexamer controls key sensing, signaling, regulation, and effector responses to DNA double-strand breaks including ATM activation, homologous recombinational repair, microhomology-mediated end joining and, in some organisms, non-homologous end joining. We propose that this is possible because each MRN subunit can exist in three or more distinct states; thus, the trimer of MRN dimers can exist in a stunning 6(3) or 216 states, a number that can be expanded further when post-translational modifications are taken into account. MRN can therefore be considered as a molecular computer that effectively assesses optimal responses and pathway choice based upon its states as set by cell status and the nature of the DNA damage. This extreme multi-state concept demands a paradigm shift from striving to understand DNA damage responses in separate terms of signaling, checkpoint, and effector proteins: we must now endeavor to characterize conformational and assembly states of MRN and other DNA repair machines that couple, coordinate, and control biological outcomes. Addressing the emerging challenge of gaining a detailed molecular understanding of MRN and other multi-state dynamic DNA repair machines promises to provide opportunities to develop master keys for controlling cell biology with probable impacts on therapeutic interventions.
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures


References
-
- Hopfner KP, Putnam CD, Tainer J. DNA double-strand break repair from head to tail. Current Opinion in Structural Biology. 2002;12:115–122. - PubMed
-
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. - PubMed
-
- Sun H, Treco D, Schultes NP, Szostak JW. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

