PER2 controls lipid metabolism by direct regulation of PPARγ
- PMID: 21035761
- PMCID: PMC4103168
- DOI: 10.1016/j.cmet.2010.10.005
PER2 controls lipid metabolism by direct regulation of PPARγ
Abstract
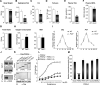
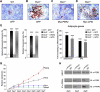
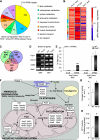
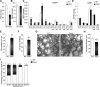
Accumulating evidence highlights intriguing interplays between circadian and metabolic pathways. We show that PER2 directly and specifically represses PPARγ, a nuclear receptor critical in adipogenesis, insulin sensitivity, and inflammatory response. PER2-deficient mice display altered lipid metabolism with drastic reduction of total triacylglycerol and nonesterified fatty acids. PER2 exerts its inhibitory function by blocking PPARγ recruitment to target promoters and thereby transcriptional activation. Whole-genome microarray profiling demonstrates that PER2 dictates the specificity of PPARγ transcriptional activity. Indeed, lack of PER2 results in enhanced adipocyte differentiation of cultured fibroblasts. PER2 targets S112 in PPARγ, a residue whose mutation has been associated with altered lipid metabolism. Lipidomic profiling demonstrates that PER2 is necessary for normal lipid metabolism in white adipocyte tissue. Our findings support a scenario in which PER2 controls the proadipogenic activity of PPARγ by operating as its natural modulator, thereby revealing potential avenues of pharmacological and therapeutic intervention.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

References
-
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. - PubMed
-
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. - PubMed
-
- Binas B, Han XX, Erol E, Luiken JJ, Glatz JF, Dyck DJ, Motazavi R, Adihetty PJ, Hood DA, Bonen A. A null mutation in H-FABP only partially inhibits skeletal muscle fatty acid metabolism. American journal of physiology. 2003;285:E481–489. - PubMed
-
- Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

