Control of eukaryotic gene expression: gene loops and transcriptional memory
- PMID: 21036187
- PMCID: PMC3305805
- DOI: 10.1016/j.advenzreg.2010.10.001
Control of eukaryotic gene expression: gene loops and transcriptional memory
Abstract
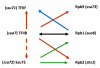
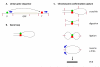
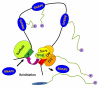
Gene loops are dynamic structures that juxtapose promoter–terminator regions of Pol II-transcribed genes. Although first described in yeast, gene loops have now been identified in yeast and mammalian cells. Looping requires components of the transcription preinitiation complex, the pre-mRNA 30-end processing machinery, and subunits of the nuclear pore complex. Loop formation is transcription-dependent, but neither basal nor activated transcription requires looping. Rather, looping appears to affect cellular memory of recent transcriptional activity, enabling a more rapid response to subsequent stimuli. The nuclear pore has been implicated in both memory and looping. Our working model is that loops are formed and/or maintained at the nuclear pore to facilitate hand-off of Pol II form the terminator to the promoter, thereby bypassing Pol II recruitment as the rate-limiting step in reactivation of transcription. Involvement of the nuclear pore also suggests that looping might facilitate mRNA export to the cytoplasm. The technology now exists to test these ideas.
Figures
References
-
- Berroteran RW, Ware DE, Hampsey M. The sua8 suppressors of Saccharomyces cerevisiae encode replacements of conserved residues within the largest subunit of RNA polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutations. Mol Cell Biol. 1994;14:226–237. - PMC - PubMed
-
- Chambeyron S, Bickmore WA. Does looping and clustering in the nucleus regulate gene expression? Curr Opin Cell Biol. 2004;16:256–262. - PubMed
-
- Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

