Methods for transcriptomic analyses of the porcine host immune response: application to Salmonella infection using microarrays
- PMID: 21036404
- PMCID: PMC6545292
- DOI: 10.1016/j.vetimm.2010.10.006
Methods for transcriptomic analyses of the porcine host immune response: application to Salmonella infection using microarrays
Abstract
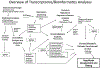
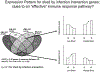
Technological developments in both the collection and analysis of molecular genetic data over the past few years have provided new opportunities for an improved understanding of the global response to pathogen exposure. Such developments are particularly dramatic for scientists studying the pig, where tools to measure the expression of tens of thousands of transcripts, as well as unprecedented data on the porcine genome sequence, have combined to expand our abilities to elucidate the porcine immune system. In this review, we describe these recent developments in the context of our work using primarily microarrays to explore gene expression changes during infection of pigs by Salmonella. Thus while the focus is not a comprehensive review of all possible approaches, we provide links and information on both the tools we use as well as alternatives commonly available for transcriptomic data collection and analysis of porcine immune responses. Through this review, we expect readers will gain an appreciation for the necessary steps to plan, conduct, analyze and interpret the data from transcriptomic analyses directly applicable to their research interests.
Copyright © 2010 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest statement
The authors declare there are no conflicts of interest to be disclosed. Funding of several aspects of the work described in this manuscript performed in the authors’ laboratories came from the USDA-NRICGP, Iowa State University Center for Integrated Animal Genomics, USDA-Food Safety Consortium, USDA-ARS, and the National Pork Board. None of these study sponsors had a role in writing or submission of this manuscript.
Figures

References
-
- Bates JS, Petry DB, Eudy J, Bough L, Johnson RK, 2008. Differential expression in lung and bronchial lymph node of pigs with high and low responses to infection with porcine reproductive and respiratory syndrome virus. J. Anim. Sci 86, 3279–3289. - PubMed
-
- Beck GC, Rafat N, Brinkkoetter P, Hanusch C, Schulte J, Haak M, van Ackern K, van der Woude FJ, Yard BA, 2006. Heterogeneity in lipopolysaccharide responsiveness of endothelial cells identified by gene expression profiling: role of transcription factors. Clin. Exp. Immunol 143, 523–533. - PMC - PubMed
-
- Belacel N, Wang Q, Cuperlovic-Culf M, 2006. Clustering methods for microarray gene expression data. OMICS 10, 507–531. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

