Intermediary metabolism in protists: a sequence-based view of facultative anaerobic metabolism in evolutionarily diverse eukaryotes
- PMID: 21036663
- PMCID: PMC3021972
- DOI: 10.1016/j.protis.2010.09.001
Intermediary metabolism in protists: a sequence-based view of facultative anaerobic metabolism in evolutionarily diverse eukaryotes
Abstract
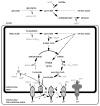
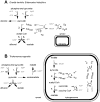
Protists account for the bulk of eukaryotic diversity. Through studies of gene and especially genome sequences the molecular basis for this diversity can be determined. Evident from genome sequencing are examples of versatile metabolism that go far beyond the canonical pathways described for eukaryotes in textbooks. In the last 2-3 years, genome sequencing and transcript profiling has unveiled several examples of heterotrophic and phototrophic protists that are unexpectedly well-equipped for ATP production using a facultative anaerobic metabolism, including some protists that can (Chlamydomonas reinhardtii) or are predicted (Naegleria gruberi, Acanthamoeba castellanii, Amoebidium parasiticum) to produce H(2) in their metabolism. It is possible that some enzymes of anaerobic metabolism were acquired and distributed among eukaryotes by lateral transfer, but it is also likely that the common ancestor of eukaryotes already had far more metabolic versatility than was widely thought a few years ago. The discussion of core energy metabolism in unicellular eukaryotes is the subject of this review. Since genomic sequencing has so far only touched the surface of protist diversity, it is anticipated that sequences of additional protists may reveal an even wider range of metabolic capabilities, while simultaneously enriching our understanding of the early evolution of eukaryotes.
Copyright © 2010 Elsevier GmbH. All rights reserved.
Figures


Similar articles
-
The genome of Naegleria gruberi illuminates early eukaryotic versatility.Cell. 2010 Mar 5;140(5):631-42. doi: 10.1016/j.cell.2010.01.032. Cell. 2010. PMID: 20211133
-
Biochemistry and evolution of anaerobic energy metabolism in eukaryotes.Microbiol Mol Biol Rev. 2012 Jun;76(2):444-95. doi: 10.1128/MMBR.05024-11. Microbiol Mol Biol Rev. 2012. PMID: 22688819 Free PMC article. Review.
-
Phylogenetic distributions and histories of proteins involved in anaerobic pyruvate metabolism in eukaryotes.Mol Biol Evol. 2010 Feb;27(2):311-24. doi: 10.1093/molbev/msp237. Epub 2009 Oct 5. Mol Biol Evol. 2010. PMID: 19805439
-
Anaerobic energy metabolism in unicellular photosynthetic eukaryotes.Biochim Biophys Acta. 2013 Feb;1827(2):210-23. doi: 10.1016/j.bbabio.2012.08.002. Epub 2012 Aug 10. Biochim Biophys Acta. 2013. PMID: 22902601 Review.
-
Arginine deiminase pathway enzymes: evolutionary history in metamonads and other eukaryotes.BMC Evol Biol. 2016 Oct 6;16(1):197. doi: 10.1186/s12862-016-0771-4. BMC Evol Biol. 2016. PMID: 27716026 Free PMC article.
Cited by
-
Oxygen suppression of macroscopic multicellularity.Nat Commun. 2021 May 14;12(1):2838. doi: 10.1038/s41467-021-23104-0. Nat Commun. 2021. PMID: 33990594 Free PMC article.
-
Response of the ubiquitous pelagic diatom Thalassiosira weissflogii to darkness and anoxia.PLoS One. 2013 Dec 2;8(12):e82605. doi: 10.1371/journal.pone.0082605. eCollection 2013. PLoS One. 2013. PMID: 24312664 Free PMC article.
-
Regulation of Microalgal Photosynthetic Electron Transfer.Plants (Basel). 2024 Jul 29;13(15):2103. doi: 10.3390/plants13152103. Plants (Basel). 2024. PMID: 39124221 Free PMC article. Review.
-
Adaptation to life on land at high O2 via transition from ferredoxin-to NADH-dependent redox balance.Proc Biol Sci. 2019 Aug 28;286(1909):20191491. doi: 10.1098/rspb.2019.1491. Epub 2019 Aug 21. Proc Biol Sci. 2019. PMID: 31431166 Free PMC article.
-
The Oxymonad Genome Displays Canonical Eukaryotic Complexity in the Absence of a Mitochondrion.Mol Biol Evol. 2019 Oct 1;36(10):2292-2312. doi: 10.1093/molbev/msz147. Mol Biol Evol. 2019. PMID: 31387118 Free PMC article.
References
-
- Abe T, Hoshino T, Nakamura A, Takaya N. Anaerobic elemental sulfur reduction by fungus Fusarium oxysporum. Biosci Biotechnol Biochem. 2007;71:2402–2407. - PubMed
-
- Allen JW, Ferguson SJ, Ginger ML. Distinctive biochemistry in the trypanosome mitochondrial intermembrane space suggests a model for stepwise evolution of the MIA pathway for import of cysteine-rich proteins. FEBS Lett. 2008b;582:2817–2825. - PubMed
-
- Amaral Zettler LA, Gomez F, Zettler E, Keenan BG, Amils R, Sogin ML. Microbiology: eukaryotic diversity in Spain’s River of Fire. Nature. 2002;417:137. - PubMed
-
- Anantharaman V, Iyer LM, Aravind L. Comparative genomics of protists: new insights into the evolution of eukaryotic signal transduction and gene regulation. Annu Rev Microbiol. 2007;61:453–475. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

