The high fidelity and unique error signature of human DNA polymerase epsilon
- PMID: 21036870
- PMCID: PMC3061053
- DOI: 10.1093/nar/gkq1034
The high fidelity and unique error signature of human DNA polymerase epsilon
Abstract
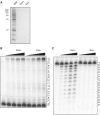
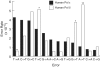
Bulk replicative DNA synthesis in eukaryotes is highly accurate and efficient, primarily because of two DNA polymerases (Pols): Pols δ and ε. The high fidelity of these enzymes is due to their intrinsic base selectivity and proofreading exonuclease activity which, when coupled with post-replication mismatch repair, helps to maintain human mutation rates at less than one mutation per genome duplication. Conditions that reduce polymerase fidelity result in increased mutagenesis and can lead to cancer in mice. Whereas yeast Pol ε has been well characterized, human Pol ε remains poorly understood. Here, we present the first report on the fidelity of human Pol ε. We find that human Pol ε carries out DNA synthesis with high fidelity, even in the absence of its 3'→5' exonucleolytic proofreading and is significantly more accurate than yeast Pol ε. Though its spectrum of errors is similar to that of yeast Pol ε, there are several notable exceptions. These include a preference of the human enzyme for T→A over A→T transversions. As compared with other replicative DNA polymerases, human Pol ε is particularly accurate when copying homonucleotide runs of 4-5 bases. The base pair substitution specificity and high fidelity for frameshift errors observed for human Pol ε are distinct from the errors made by human Pol δ.
Figures


Similar articles
-
Fidelity of DNA polymerase epsilon holoenzyme from budding yeast Saccharomyces cerevisiae.J Biol Chem. 2002 Oct 4;277(40):37422-9. doi: 10.1074/jbc.M204476200. Epub 2002 Jul 17. J Biol Chem. 2002. PMID: 12124389
-
Unique error signature of the four-subunit yeast DNA polymerase epsilon.J Biol Chem. 2003 Oct 31;278(44):43770-80. doi: 10.1074/jbc.M306893200. Epub 2003 Jul 25. J Biol Chem. 2003. PMID: 12882968
-
Fidelity of mammalian DNA replication and replicative DNA polymerases.Biochemistry. 1991 Dec 24;30(51):11751-9. doi: 10.1021/bi00115a003. Biochemistry. 1991. PMID: 1751492
-
Extrinsic proofreading.DNA Repair (Amst). 2022 Sep;117:103369. doi: 10.1016/j.dnarep.2022.103369. Epub 2022 Jul 4. DNA Repair (Amst). 2022. PMID: 35850061 Free PMC article. Review.
-
A panoply of errors: polymerase proofreading domain mutations in cancer.Nat Rev Cancer. 2016 Feb;16(2):71-81. doi: 10.1038/nrc.2015.12. Nat Rev Cancer. 2016. PMID: 26822575 Review.
Cited by
-
Characterization of hotspot exonuclease domain mutations in the DNA polymerase ϵ gene in endometrial cancer.Front Oncol. 2022 Oct 12;12:1018034. doi: 10.3389/fonc.2022.1018034. eCollection 2022. Front Oncol. 2022. PMID: 36313640 Free PMC article.
-
Discovery, characterisation and genomic variation of six novel Gammapapillomavirus types from penile swabs in South Africa.Papillomavirus Res. 2019 Jun;7:102-111. doi: 10.1016/j.pvr.2019.02.005. Epub 2019 Mar 4. Papillomavirus Res. 2019. PMID: 30844514 Free PMC article.
-
Replicative DNA polymerase defects in human cancers: Consequences, mechanisms, and implications for therapy.DNA Repair (Amst). 2017 Aug;56:16-25. doi: 10.1016/j.dnarep.2017.06.003. Epub 2017 Jun 9. DNA Repair (Amst). 2017. PMID: 28687338 Free PMC article. Review.
-
On the sequence-directed nature of human gene mutation: the role of genomic architecture and the local DNA sequence environment in mediating gene mutations underlying human inherited disease.Hum Mutat. 2011 Oct;32(10):1075-99. doi: 10.1002/humu.21557. Epub 2011 Sep 2. Hum Mutat. 2011. PMID: 21853507 Free PMC article. Review.
-
Eukaryotic genome instability in light of asymmetric DNA replication.Crit Rev Biochem Mol Biol. 2016;51(1):43-52. doi: 10.3109/10409238.2015.1117055. Epub 2015 Dec 20. Crit Rev Biochem Mol Biol. 2016. PMID: 26822554 Free PMC article. Review.
References
-
- Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. - PubMed
-
- Karthikeyan R, Vonarx EJ, Straffon AF, Simon M, Faye G, Kunz BA. Evidence from mutational specificity studies that yeast DNA polymerases delta and epsilon replicate different DNA strands at an intracellular replication fork. J. Mol. Biol. 2000;299:405–419. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

