Chlamydia-specific CD4 T cell clones control Chlamydia muridarum replication in epithelial cells by nitric oxide-dependent and -independent mechanisms
- PMID: 21037093
- PMCID: PMC3073083
- DOI: 10.4049/jimmunol.1002596
Chlamydia-specific CD4 T cell clones control Chlamydia muridarum replication in epithelial cells by nitric oxide-dependent and -independent mechanisms
Abstract
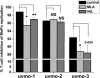
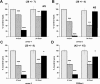
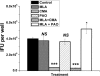
Chlamydia trachomatis serovars D-K are sexually transmitted intracellular bacterial pathogens that replicate in epithelial cells lining the human reproductive tract. It is clear from knockout mice and T cell depletion studies using Chlamydia muridarum that MHC class II and CD4 T cells are critical for clearing bacteria from the murine genital tract. It is not clear how CD4 T cells interact with infected epithelial cells to mediate bacterial clearance in vivo. Previous work using an epithelial tumor cell line showed that a Chlamydia-specific CD4 T cell clone was able to inhibit C. muridarum replication in vitro via induction of epithelial NO production. We have previously shown that Chlamydia-specific CD4 T cell clones can recognize and be activated by infected reproductive tract epithelial cells and block Chlamydia replication in them. We extend those observations by investigating the mechanism used by a panel of CD4 T cell clones to control Chlamydia replication in epithelial cells. We found that Chlamydia-specific CD4 T cell clones were cytolytic, but that cytolysis was not likely critical for controlling C. muridarum replication. For one, CD4 T cell clone-induced epithelial NO production was critical for controlling replication; however, the most potent CD4 T cell clones were dependent on T cell degranulation for replication control with only a minor additional contribution from NO production. We discuss our data as they relate to existing knockout mouse studies addressing mechanisms of T cell-mediated control of Chlamydia replication and their implications for intracellular epithelial pathogens in mouse models.
Figures
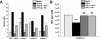





Similar articles
-
Perforin is detrimental to controlling [corrected] C. muridarum replication in vitro, but not in vivo.PLoS One. 2013 May 14;8(5):e63340. doi: 10.1371/journal.pone.0063340. Print 2013. PLoS One. 2013. PMID: 23691028 Free PMC article.
-
Plac8-dependent and inducible NO synthase-dependent mechanisms clear Chlamydia muridarum infections from the genital tract.J Immunol. 2012 Feb 15;188(4):1896-904. doi: 10.4049/jimmunol.1102764. Epub 2012 Jan 11. J Immunol. 2012. PMID: 22238459 Free PMC article.
-
Chlamydia muridarum-specific CD4 T-cell clones recognize infected reproductive tract epithelial cells in an interferon-dependent fashion.Infect Immun. 2009 Oct;77(10):4469-79. doi: 10.1128/IAI.00491-09. Epub 2009 Aug 10. Infect Immun. 2009. PMID: 19667042 Free PMC article.
-
T cell responses to Chlamydia.Pathog Dis. 2021 Mar 31;79(4):ftab014. doi: 10.1093/femspd/ftab014. Pathog Dis. 2021. PMID: 33693620 Free PMC article. Review.
-
Chlamydia Spreading from the Genital Tract to the Gastrointestinal Tract - A Two-Hit Hypothesis.Trends Microbiol. 2018 Jul;26(7):611-623. doi: 10.1016/j.tim.2017.12.002. Epub 2017 Dec 27. Trends Microbiol. 2018. PMID: 29289422 Free PMC article. Review.
Cited by
-
Evaluation of a multisubunit recombinant polymorphic membrane protein and major outer membrane protein T cell vaccine against Chlamydia muridarum genital infection in three strains of mice.Vaccine. 2014 Aug 6;32(36):4672-80. doi: 10.1016/j.vaccine.2014.06.002. Epub 2014 Jun 30. Vaccine. 2014. PMID: 24992718 Free PMC article.
-
Modeling the transcriptome of genital tract epithelial cells and macrophages in healthy mucosa versus mucosa inflamed by Chlamydia muridarum infection.Pathog Dis. 2015 Dec;73(9):ftv100. doi: 10.1093/femspd/ftv100. Epub 2015 Oct 29. Pathog Dis. 2015. PMID: 26519447 Free PMC article.
-
Perforin is detrimental to controlling [corrected] C. muridarum replication in vitro, but not in vivo.PLoS One. 2013 May 14;8(5):e63340. doi: 10.1371/journal.pone.0063340. Print 2013. PLoS One. 2013. PMID: 23691028 Free PMC article.
-
BHLHE40 drives protective polyfunctional CD4 T cell differentiation in the female reproductive tract against Chlamydia.PLoS Pathog. 2024 Jan 25;20(1):e1011983. doi: 10.1371/journal.ppat.1011983. eCollection 2024 Jan. PLoS Pathog. 2024. PMID: 38271477 Free PMC article.
-
Integrin α4β1 is necessary for CD4+ T cell-mediated protection against genital Chlamydia trachomatis infection.J Immunol. 2014 May 1;192(9):4284-93. doi: 10.4049/jimmunol.1303238. Epub 2014 Mar 21. J Immunol. 2014. PMID: 24659687 Free PMC article.
References
-
- CDC, editor. Sexually Transmitted Disease Surveillance, 2008. U.S. Department of Health and Human Services; Atlanta, GA: 2009.
-
- Brunham RC, Pourbohloul B, Mak S, White R, Rekart ML. The Unexpected Impact of a Chlamydia trachomatis Infection Control Program on Susceptibility to Reinfection. J Infect Dis. 2005;192:1836–1844. - PubMed
-
- Haggerty CL, Gottlieb SL, Taylor BD, Low N, Xu F, Ness RB. Risk of sequelae after Chlamydia trachomatis genital infection in women. J Infect Dis. 201(Suppl 2):S134–155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

