Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development
- PMID: 21037105
- PMCID: PMC2990135
- DOI: 10.1105/tpc.110.079962
Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development
Abstract
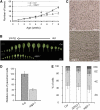
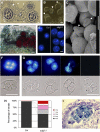
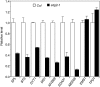
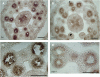
Histone H3 lysine 4 trimethylation (H3K4me3) is abundant in euchromatin and is in general associated with transcriptional activation in eukaryotes. Although some Arabidopsis thaliana SET DOMAIN GROUP (SDG) genes have been previously shown to be involved in H3K4 methylation, they are unlikely to be responsible for global genome-wide deposition of H3K4me3. Most strikingly, sparse knowledge is currently available about the role of histone methylation in gametophyte development. In this study, we show that the previously uncharacterized SDG2 is required for global H3K4me3 deposition and its loss of function causes wide-ranging defects in both sporophyte and gametophyte development. Transcriptome analyses of young flower buds have identified 452 genes downregulated by more than twofold in the sdg2-1 mutant; among them, 11 genes, including SPOROCYTELESS/NOZZLE (SPL/NZZ) and MALE STERILITY1 (MS1), have been previously shown to be essential for male and/or female gametophyte development. We show that both SPL/NZZ and MS1 contain bivalent chromatin domains enriched simultaneously with the transcriptionally active mark H3K4me3 and the transcriptionally repressive mark H3K27me3 and that SDG2 is specifically required for the H3K4me3 deposition. Our data suggest that SDG2-mediated H3K4me3 deposition poises SPL/NZZ and MS1 for transcriptional activation, forming a key regulatory mechanism in the gene networks responsible for gametophyte development.
Figures




References
-
- Aarts M.G., Hodge R., Kalantidis K., Florack D., Wilson Z.A., Mulligan B.J., Stiekema W.J., Scott R., Pereira A. (1997). The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12: 615–623 - PubMed
-
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 - PubMed
-
- Alvarez-Venegas R., Pien S., Sadder M., Witmer X., Grossniklaus U., Avramova Z. (2003). ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 13: 627–637 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

