Expanding the chemical scope of RNA:methyltransferases to site-specific alkynylation of RNA for click labeling
- PMID: 21037259
- PMCID: PMC3061074
- DOI: 10.1093/nar/gkq825
Expanding the chemical scope of RNA:methyltransferases to site-specific alkynylation of RNA for click labeling
Abstract
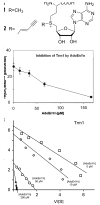
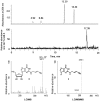
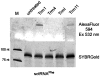
This work identifies the combination of enzymatic transfer and click labeling as an efficient method for the site-specific tagging of RNA molecules for biophysical studies. A double-activated analog of the ubiquitous co-substrate S-adenosyl-l-methionine was employed to enzymatically transfer a five carbon chain containing a terminal alkynyl moiety onto RNA. The tRNA:methyltransferase Trm1 transferred the extended alkynyl moiety to its natural target, the N2 of guanosine 26 in tRNA(Phe). LC/MS and LC/MS/MS techniques were used to detect and characterize the modified nucleoside as well as its cycloaddition product with a fluorescent azide. The latter resulted from a labeling reaction via Cu(I)-catalyzed azide-alkyne 1,3-cycloaddition click chemistry, producing site-specifically labeled RNA whose suitability for single molecule fluorescence experiments was verified in fluorescence correlation spectroscopy experiments.
Figures



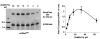

Similar articles
-
Fluorescent DNA labeling by N-mustard analogues of S-adenosyl-L-methionine.Chembiochem. 2012 Oct 15;13(15):2225-33. doi: 10.1002/cbic.201200438. Epub 2012 Sep 7. Chembiochem. 2012. PMID: 22961989
-
Programmable sequence-specific click-labeling of RNA using archaeal box C/D RNP methyltransferases.Nucleic Acids Res. 2012 Aug;40(14):6765-73. doi: 10.1093/nar/gks381. Epub 2012 May 7. Nucleic Acids Res. 2012. PMID: 22564896 Free PMC article.
-
A selenium-based click AdoMet analogue for versatile substrate labeling with wild-type protein methyltransferases.Chembiochem. 2012 May 29;13(8):1167-73. doi: 10.1002/cbic.201100781. Epub 2012 Apr 30. Chembiochem. 2012. PMID: 22549896
-
Enzymatic formation of N2,N2-dimethylguanosine in eukaryotic tRNA: importance of the tRNA architecture.Biochimie. 1995;77(1-2):54-61. doi: 10.1016/0300-9084(96)88104-1. Biochimie. 1995. PMID: 7599276 Review.
-
tRNA-m1G methyltransferase interactions: touching bases with structure.Biochimie. 1995;77(1-2):62-5. doi: 10.1016/0300-9084(96)88105-3. Biochimie. 1995. PMID: 7599277 Review.
Cited by
-
Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.Nucleic Acids Res. 2015 May 19;43(9):4602-13. doi: 10.1093/nar/gkv206. Epub 2015 Apr 8. Nucleic Acids Res. 2015. PMID: 25855808 Free PMC article.
-
Quantum Chemical Calculations and Experimental Validation of the Photoclick Reaction for Fluorescent Labeling of the 5' cap of Eukaryotic mRNAs.ChemistryOpen. 2015 Jun;4(3):295-301. doi: 10.1002/open.201402104. Epub 2015 Feb 4. ChemistryOpen. 2015. PMID: 26246991 Free PMC article.
-
Site-specific terminal and internal labeling of RNA by poly(A) polymerase tailing and copper-catalyzed or copper-free strain-promoted click chemistry.Nucleic Acids Res. 2012 May;40(10):e78. doi: 10.1093/nar/gks062. Epub 2012 Feb 16. Nucleic Acids Res. 2012. PMID: 22344697 Free PMC article.
-
Nicking enzyme-based internal labeling of DNA at multiple loci.Nat Protoc. 2012 Mar 8;7(4):643-53. doi: 10.1038/nprot.2012.008. Nat Protoc. 2012. PMID: 22402634
-
Functional integration of a semi-synthetic azido-queuosine derivative into translation and a tRNA modification circuit.Nucleic Acids Res. 2022 Oct 14;50(18):10785-10800. doi: 10.1093/nar/gkac822. Nucleic Acids Res. 2022. PMID: 36169220 Free PMC article.
References
-
- Bujnicki JM, Droogmans L, Grosjean H, Purushothaman SK, Lapeyre B. Bioinformatics-guided identification and experimental characterization of novel RNA methyltransferases. Nucleic Acids Mol. Biol. 2004;15:139–168.
-
- Michelot R, Legreverend M, Farrugia G, Lederer E. New studies on inhibition of tRNA N2 guanine methyltransferase by S-adenosyl-homocysteine and S-adenosyl-methionine analogs. Biochimie. 1976;58:201–205. - PubMed
-
- Leboy PS, Glick JM, Steiner FG, Haney S, Borchardt RT. S-adenosylhomocysteine analogues as inhibitors of specific tRNA methylation. Biochim. Biophys. Acta. 1978;520:153–163. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

