starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data
- PMID: 21037263
- PMCID: PMC3013664
- DOI: 10.1093/nar/gkq1056
starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data
Abstract
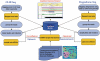
MicroRNAs (miRNAs) represent an important class of small non-coding RNAs (sRNAs) that regulate gene expression by targeting messenger RNAs. However, assigning miRNAs to their regulatory target genes remains technically challenging. Recently, high-throughput CLIP-Seq and degradome sequencing (Degradome-Seq) methods have been applied to identify the sites of Argonaute interaction and miRNA cleavage sites, respectively. In this study, we introduce a novel database, starBase (sRNA target Base), which we have developed to facilitate the comprehensive exploration of miRNA-target interaction maps from CLIP-Seq and Degradome-Seq data. The current version includes high-throughput sequencing data generated from 21 CLIP-Seq and 10 Degradome-Seq experiments from six organisms. By analyzing millions of mapped CLIP-Seq and Degradome-Seq reads, we identified ∼1 million Ago-binding clusters and ∼2 million cleaved target clusters in animals and plants, respectively. Analyses of these clusters, and of target sites predicted by 6 miRNA target prediction programs, resulted in our identification of approximately 400,000 and approximately 66,000 miRNA-target regulatory relationships from CLIP-Seq and Degradome-Seq data, respectively. Furthermore, two web servers were provided to discover novel miRNA target sites from CLIP-Seq and Degradome-Seq data. Our web implementation supports diverse query types and exploration of common targets, gene ontologies and pathways. The starBase is available at http://starbase.sysu.edu.cn/.
Figures

References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. - PubMed
-
- Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl.):S8–13. - PubMed
-
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

