Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors sigmaB, sigmaC, sigmaH, and sigmaL in Listeria monocytogenes
- PMID: 21037293
- PMCID: PMC3019704
- DOI: 10.1128/AEM.00952-10
Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors sigmaB, sigmaC, sigmaH, and sigmaL in Listeria monocytogenes
Abstract
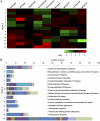
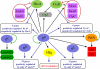
A set of seven Listeria monocytogenes 10403S mutant strains, each bearing an in-frame null mutation in a gene encoding a key regulatory protein, was used to characterize transcriptional networks in L. monocytogenes; the seven regulatory proteins addressed include all four L. monocytogenes alternative sigma factors (σ(B), σ(C), σ(H), and σ(L)), the virulence gene regulator PrfA, and the heat shock-related negative regulators CtsR and HrcA. Whole-genome microarray analyses, used to identify regulons for each of these 7 transcriptional regulators, showed considerable overlap among regulons. Among 188 genes controlled by more than one regulator, 176 were coregulated by σ(B), including 92 genes regulated by both σ(B) and σ(H) (with 18 of these genes coregulated by σ(B), σ(H), and at least one additional regulator) and 31 genes regulated by both σ(B) and σ(L) (with 10 of these genes coregulated by σ(B), σ(L), and at least one additional regulator). Comparative phenotypic characterization measuring acid resistance, heat resistance, intracellular growth in J774 cells, invasion into Caco-2 epithelial cells, and virulence in the guinea pig model indicated contributions of (i) σ(B) to acid resistance, (ii) CtsR to heat resistance, and (iii) PrfA, σ(B), and CtsR to virulence-associated characteristics. Loss of the remaining transcriptional regulators (i.e., sigH, sigL, or sigC) resulted in limited phenotypic consequences associated with stress survival and virulence. Identification of overlaps among the regulons provides strong evidence supporting the existence of complex regulatory networks that appear to provide the cell with regulatory redundancies, along with the ability to fine-tune gene expression in response to rapidly changing environmental conditions.
Figures

References
-
- Arous, S., C. Buchrieser, P. Folio, P. Glaser, A. Namane, M. Hebraud, and Y. Hechard. 2004. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology 150:1581-1590. - PubMed
-
- Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

