Identifying the combinatorial effects of histone modifications by association rule mining in yeast
- PMID: 21037963
- PMCID: PMC2964047
- DOI: 10.4137/ebo.s5602
Identifying the combinatorial effects of histone modifications by association rule mining in yeast
Abstract
Eukaryotic genomes are packaged into chromatin by histone proteins whose chemical modification can profoundly influence gene expression. The histone modifications often act in combinations, which exert different effects on gene expression. Although a number of experimental techniques and data analysis methods have been developed to study histone modifications, it is still very difficult to identify the relationships among histone modifications on a genome-wide scale.We proposed a method to identify the combinatorial effects of histone modifications by association rule mining. The method first identified Functional Modification Transactions (FMTs) and then employed association rule mining algorithm and statistics methods to identify histone modification patterns. We applied the proposed methodology to Pokholok et al's data with eight sets of histone modifications and Kurdistani et al's data with eleven histone acetylation sites. Our method succeeds in revealing two different global views of histone modification landscapes on two datasets and identifying a number of modification patterns some of which are supported by previous studies.We concentrate on combinatorial effects of histone modifications which significantly affect gene expression. Our method succeeds in identifying known interactions among histone modifications and uncovering many previously unknown patterns. After in-depth analysis of possible mechanism by which histone modification patterns can alter transcriptional states, we infer three possible modification pattern reading mechanism ('redundant', 'trivial', 'dominative'). Our results demonstrate several histone modification patterns which show significant correspondence between yeast and human cells.
Keywords: association rule; histone code; yeast.
Figures

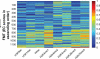

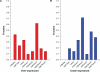

References
-
- Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002 Dec 13;111(6):771–8. - PubMed
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007 May 24;447(7143):407–12. - PubMed
-
- Rando OJ. Global patterns of histone modifications. Curr Opin Genet Dev. 2007 Apr;17(2):94–9. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007 Feb 23;128(4):693–705. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

