Characterization of a novel family of fibronectin-binding proteins with M23 peptidase domains from Treponema denticola
- PMID: 21040511
- PMCID: PMC3103763
- DOI: 10.1111/j.2041-1014.2010.00584.x
Characterization of a novel family of fibronectin-binding proteins with M23 peptidase domains from Treponema denticola
Abstract
Interactions with fibronectin are important in the virulence strategies of a range of disease-related bacteria. The periodontitis-associated oral spirochaete Treponema denticola expresses at least two fibronectin-binding proteins, designated Msp (major surface protein) and OppA (oligopeptide-binding protein homologue). To identify other T. denticola outer membrane fibronectin-binding proteins, the amino acid sequence of the Treponema pallidum fibronectin-binding protein Tp0155 was used to survey the T. denticola genome. Seven T. denticola genes encoding orthologous proteins were identified. All but two were expressed in Escherichia coli and purified recombinant proteins bound fibronectin. Using antibodies to the N-terminal region of Tp0155, it was demonstrated that T. denticola TDE2318, with highest homology to Tp0155, was cell surface localized. Like Tp0155, the seven T. denticola proteins contained an M23 peptidase domain and four (TDE2318, TDE2753, TDE1738, TDE1297) contained one or two LysM domains. M23 peptidases can degrade peptidoglycan whereas LysM domains recognize carbohydrate polymers. In addition, TDE1738 may act as a bacteriocin based on homology with other bacterial lysins and the presence of an adjacent gene encoding a putative immunity factor. Collectively, these results suggest that T. denticola expresses fibronectin-binding proteins associated with the cell surface that may also have cell wall modifying or lytic functions.
© 2010 John Wiley & Sons A/S.
Figures

 ) to lowest (e.g. H). The LysM motif YxxxxGxx(Hyd) (Turner et al., 2004) is boxed. Asterisk indicates residues that do not fit the specified motif.
) to lowest (e.g. H). The LysM motif YxxxxGxx(Hyd) (Turner et al., 2004) is boxed. Asterisk indicates residues that do not fit the specified motif.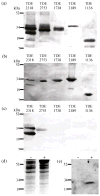
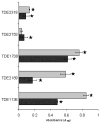

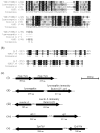
 ) to lowest (e.g. A). (C) Diagram depicting genes encoding potential and known peptidoglycan degradation enzymes and adjacent immunity factors which provide protection from autolysis by the peptidoglycanolytic enzyme. (i) The chromosomal gene TDE1738 from T. denticola encodes for a potential peptidoglycan-degrading enzyme, and TDE1739 for a potential immunity factor. (ii) Lysostaphin and adjacent lysostaphin immunity factor (Lif/Epr) are encoded on the plasmid pACK1 of S. simulans biovar staphylolyticus. (iii) The gene encoding zoocin A and corresponding zoocin A immunity factor (Zif) is on the chromosome of Streptococcus equi subsp zooepidemicus, while (iv) ale-1 and adjacent endopeptidase resistance (epr) genes are located on a plasmid in Staphylococcus capitis. (v) Tp0706 from the Treponema pallidum chromosome, with homology to TDE1738 may also be an enzyme with peptidoglycan-degrading activity and the adjacent gene Tp0705 is annotated as a pencillin-binding protein in the same manner as zoocin A immunity factor (Zif).
) to lowest (e.g. A). (C) Diagram depicting genes encoding potential and known peptidoglycan degradation enzymes and adjacent immunity factors which provide protection from autolysis by the peptidoglycanolytic enzyme. (i) The chromosomal gene TDE1738 from T. denticola encodes for a potential peptidoglycan-degrading enzyme, and TDE1739 for a potential immunity factor. (ii) Lysostaphin and adjacent lysostaphin immunity factor (Lif/Epr) are encoded on the plasmid pACK1 of S. simulans biovar staphylolyticus. (iii) The gene encoding zoocin A and corresponding zoocin A immunity factor (Zif) is on the chromosome of Streptococcus equi subsp zooepidemicus, while (iv) ale-1 and adjacent endopeptidase resistance (epr) genes are located on a plasmid in Staphylococcus capitis. (v) Tp0706 from the Treponema pallidum chromosome, with homology to TDE1738 may also be an enzyme with peptidoglycan-degrading activity and the adjacent gene Tp0705 is annotated as a pencillin-binding protein in the same manner as zoocin A immunity factor (Zif).Similar articles
-
Immunotopological Analysis of the Treponema denticola Major Surface Protein (Msp).J Bacteriol. 2018 Dec 20;201(2):e00528-18. doi: 10.1128/JB.00528-18. Print 2019 Jan 15. J Bacteriol. 2018. PMID: 30373754 Free PMC article.
-
Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405.Infect Immun. 2005 May;73(5):2891-8. doi: 10.1128/IAI.73.5.2891-2898.2005. Infect Immun. 2005. PMID: 15845495 Free PMC article.
-
Identification of a Treponema denticola OppA homologue that binds host proteins present in the subgingival environment.Infect Immun. 2000 Apr;68(4):1884-92. doi: 10.1128/IAI.68.4.1884-1892.2000. Infect Immun. 2000. PMID: 10722578 Free PMC article.
-
Treponema denticola major surface protein (Msp): a key player in periodontal pathogenicity and immune evasion.Arch Microbiol. 2025 Jan 18;207(2):36. doi: 10.1007/s00203-024-04223-w. Arch Microbiol. 2025. PMID: 39825920 Review.
-
Role of Treponema denticola in periodontal diseases.Crit Rev Oral Biol Med. 2001;12(5):399-413. doi: 10.1177/10454411010120050301. Crit Rev Oral Biol Med. 2001. PMID: 12002822 Review.
Cited by
-
Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species.BMC Genomics. 2016 Nov 7;17(1):882. doi: 10.1186/s12864-016-3224-y. BMC Genomics. 2016. PMID: 27821051 Free PMC article.
-
Genome Investigation of a Cariogenic Pathogen with Implications in Cardiovascular Diseases.Indian J Microbiol. 2019 Dec;59(4):451-459. doi: 10.1007/s12088-019-00823-z. Epub 2019 Sep 6. Indian J Microbiol. 2019. PMID: 31762508 Free PMC article.
-
Immunoproteomic analysis of proteins expressed by two related pathogens, Burkholderia multivorans and Burkholderia cenocepacia, during human infection.PLoS One. 2013 Nov 15;8(11):e80796. doi: 10.1371/journal.pone.0080796. eCollection 2013. PLoS One. 2013. PMID: 24260482 Free PMC article.
-
Fibronectin-binding protein TDE1579 affects cytotoxicity of Treponema denticola.Anaerobe. 2015 Dec;36:39-48. doi: 10.1016/j.anaerobe.2015.09.010. Epub 2015 Oct 9. Anaerobe. 2015. PMID: 26456217 Free PMC article.
-
An Overview of the Etiopathogenic Mechanisms Involved in the Expression of the Oral Microbiota.Clin Pract. 2025 Apr 11;15(4):80. doi: 10.3390/clinpract15040080. Clin Pract. 2025. PMID: 40310312 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

