All that glitters is not gold--founder effects complicate associations of flu mutations to disease severity
- PMID: 21040570
- PMCID: PMC2989967
- DOI: 10.1186/1743-422X-7-297
All that glitters is not gold--founder effects complicate associations of flu mutations to disease severity
Abstract
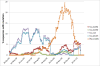
Background: The recent 2009 (H1N1) influenza A pandemic saw a rapid spread of the virus to essentially all parts of the world. In the course of its evolution, the virus acquired many mutations, some of which have been investigated in the context of increased severity due to high occurrences in fatal cases. For example, statements such as: "42.9% of individuals who died from laboratory-confirmed cases of the pandemic (H1N1) were infected with the hemagglutinin (HA) Q310 H mutant virus." give the impression that HA-Q310 H would be highly dangerous or important, while careful consideration of all available data suggests that this is unlikely to be the case.
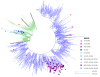
Results: We compare the mutations HA-Q310 H, PB2-K340N, HA-D239N and HA-D239G using whole genome phylogenetic trees, structural modeling in their 3 D context and complete epidemiological data from sequences to clinical outcomes. HA-Q310 H and PB2-K340N appear as isolated subtrees in the phylogenetic analysis pointing to founder effects which is consistent with their clustered temporal appearance as well as the lack of an immediate structural basis that could explain a change of phenotypes. Considering the prevailing viral genomic background, shared origin of samples (all from the city of Sao Paulo) and narrow temporal window (all death case samples within 1 month), it becomes clear that HA-Q310 H was actually a generally common mutation in the region at that time which could readily explain its increased occurrence among the few analyzed fatal cases without requiring necessarily an association with severity. In further support of this, we highlight 3 mild cases with the HA-Q310 H mutation.
Conclusions: We argue that claims of severity of any current and future flu mutation need to be critically considered in the light of phylogenetic, structural and detailed epidemiological data to distinguish increased occurrence due to possible founder effects rather than real phenotypic changes.
Figures


References
-
- Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, Kadie C, Carlson J, Yusim K, McMahon B, Gaschen B, Mallal S, Mullins JI, Nickle DC, Herbeck J, Rousseau C, Learn GH, Miura T, Brander C, Walker B, Korber B. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. - DOI - PubMed
-
- Kilander A, Rykkvin R, Dudman SG, Hungnes O. Observed association between the HA1 mutation D222G in the 2009 pandemic influenza A(H1N1) virus and severe clinical outcome, Norway 2009-2010. Euro Surveill. 2010;15 http://www.ncbi.nlm.nih.gov/pubmed/20214869 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

