A noncompetitive small molecule inhibitor of estrogen-regulated gene expression and breast cancer cell growth that enhances proteasome-dependent degradation of estrogen receptor {alpha}
- PMID: 21041310
- PMCID: PMC3009914
- DOI: 10.1074/jbc.M110.183723
A noncompetitive small molecule inhibitor of estrogen-regulated gene expression and breast cancer cell growth that enhances proteasome-dependent degradation of estrogen receptor {alpha}
Abstract
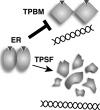
The mechanisms responsible for 17β-estradiol (E(2))-stimulated breast cancer growth and development of resistance to tamoxifen and other estrogen receptor α (ERα) antagonists are not fully understood. We describe a new tool for dissecting ERα action in breast cancer, p-fluoro-4-(1,2,3,6,-tetrahydro-1,3-dimethyl-2-oxo-6-thionpurin-8-ylthio) (TPSF), a potent small-molecule inhibitor of estrogen receptor α that does not compete with estrogen for binding to ERα. TPSF noncompetitively inhibits estrogen-dependent ERα-mediated gene expression with little inhibition of transcriptional activity by NF-κB or the androgen or glucocorticoid receptor. TPSF inhibits E(2)-ERα-mediated induction of the proteinase inhibitor 9 gene, which is activated by ERα binding to estrogen response element DNA, and the cyclin D1 gene, which is induced by tethering ERα to other DNA-bound proteins. TPSF inhibits anchorage-dependent and anchorage-independent E(2)-ERα-stimulated growth of MCF-7 cells but does not inhibit growth of ER-negative MDA-MB-231 breast cancer cells. TPSF also inhibits ERα-dependent growth in three cellular models for tamoxifen resistance; that is, 4-hydroxytamoxifen-stimulated MCF7ERαHA cells that overexpress ERα, fully tamoxifen-resistant BT474 cells that have amplified HER-2 and AIB1, and partially tamoxifen-resistant ZR-75 cells. TPSF reduces ERα protein levels in MCF-7 cells and several other cell lines without altering ERα mRNA levels. The proteasome inhibitor MG132 abolished down-regulation of ERα by TPSF. Thus, TPSF affects receptor levels at least in part due to its ability to enhance proteasome-dependent degradation of ERα. TPSF represents a novel class of ER inhibitor with significant clinical potential.
Figures

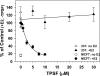
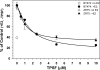
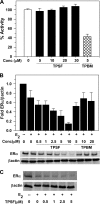
 O and fluorine substitutions at the phenyl ring. B, shown are potency and efficacy of TPSF (triangles), TPBM (squares), and 99676 (circles). Inhibition of E2-ERα activation of ERE-Luc was evaluated in dose-response studies of T47D (ERE)3-Luc cells maintained in 0.2 n
O and fluorine substitutions at the phenyl ring. B, shown are potency and efficacy of TPSF (triangles), TPBM (squares), and 99676 (circles). Inhibition of E2-ERα activation of ERE-Luc was evaluated in dose-response studies of T47D (ERE)3-Luc cells maintained in 0.2 n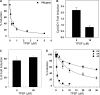



References
-
- O'Lone R., Frith M. C., Karlsson E. K., Hansen U. (2004) Mol. Endocrinol. 18, 1859–1875 - PubMed
-
- Carroll J. S., Brown M. (2006) Mol. Endocrinol. 20, 1707–1714 - PubMed
-
- Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. (2005) Cell 122, 33–43 - PubMed
-
- Jakacka M., Ito M., Weiss J., Chien P. Y., Gehm B. D., Jameson J. L. (2001) J. Biol. Chem. 276, 13615–13621 - PubMed
-
- Safe S. (2001) Vitam. Horm. 62, 231–252 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

