Proof of concept: network and systems biology approaches aid in the discovery of potent anticancer drug combinations
- PMID: 21041384
- PMCID: PMC3058926
- DOI: 10.1158/1535-7163.MCT-10-0642
Proof of concept: network and systems biology approaches aid in the discovery of potent anticancer drug combinations
Abstract
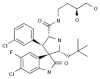
Cancer therapies that target key molecules have not fulfilled expected promises for most common malignancies. Major challenges include the incomplete understanding and validation of these targets in patients, the multiplicity and complexity of genetic and epigenetic changes in the majority of cancers, and the redundancies and cross-talk found in key signaling pathways. Collectively, the uses of single-pathway targeted approaches are not effective therapies for human malignancies. To overcome these barriers, it is important to understand the molecular cross-talk among key signaling pathways and how they may be altered by targeted agents. Innovative approaches are needed, such as understanding the global physiologic environment of target proteins and the effects of modifying them without losing key molecular details. Such strategies will aid the design of novel therapeutics and their combinations against multifaceted diseases, in which efficacious combination therapies will focus on altering multiple pathways rather than single proteins. Integrated network modeling and systems biology have emerged as powerful tools benefiting our understanding of drug mechanisms of action in real time. This review highlights the significance of the network and systems biology-based strategy and presents a proof of concept recently validated in our laboratory using the example of a combination treatment of oxaliplatin and the MDM2 inhibitor MI-219 in genetically complex and incurable pancreatic adenocarcinoma.
©2010 AACR.
Conflict of interest statement
Figures




References
-
- Wilson TR, Johnston PG, Longley DB. Anti-apoptotic mechanisms of drug resistance in cancer. Curr Cancer Drug Targets. 2009;9:307–19. - PubMed
-
- Newell DR. How to develop a successful cancer drug--molecules to medicines or targets to treatments? Eur J Cancer. 2005;41:676–82. - PubMed
-
- Heng HH, Bremer SW, Stevens JB, Ye KJ, Liu G, Ye CJ. Genetic and epigenetic heterogeneity in cancer: a genome-centric perspective. J Cell Physiol. 2009;220:538–47. - PubMed
-
- Le TC, Stathis A, Vidal L, Moore MJ, Siu LL. Choice of starting dose for molecularly targeted agents evaluated in first-in-human phase I cancer clinical trials. J Clin Oncol. 2010;28:1401–7. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

