UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway
- PMID: 21041483
- PMCID: PMC3019843
- DOI: 10.1128/MCB.00818-10
UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway
Abstract
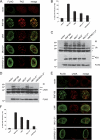
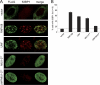
Ubiquitination regulates important cellular processes, including the DNA damage response (DDR) and DNA repair. The complexity of the ubiquitin-mediated signals is decoded by ubiquitin receptors, which contain protein modules named ubiquitin binding domains (UBDs). We previously identified a new ubiquitin ligase, RNF168, involved in DDR and endowed with two UBDs named MIU (motif interacting with ubiquitin). Here we have provided the identification of a novel UBD, the UMI (UIM- and MIU-related UBD), present in RNF168, and characterized the interaction surface with ubiquitin, centered on two Leu residues. We have demonstrated that integrity of the UMI, in addition to the MIUs, is necessary for the proper localization of RNF168 and for ubiquitination of nuclear proteins, including histone H2A. Finally, we have shown that simultaneous inactivation of UMI and MIUs prevents the recruitment to DDR foci of the crucial downstream mediator 53BP1.
Figures



References
-
- Acconcia, F., S. Sigismund, and S. Polo. 2009. Ubiquitin in trafficking: the network at work. Exp. Cell Res. 315:1610-1618. - PubMed
-
- Bergink, S., and S. Jentsch. 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458:461-467. - PubMed
-
- Bhoj, V. G., and Z. J. Chen. 2009. Ubiquitylation in innate and adaptive immunity. Nature 458:430-437. - PubMed
-
- Chen, Z. J., and L. J. Sun. 2009. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33:275-286. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
