Quantitative PCR-based competitive index for high-throughput screening of Salmonella virulence factors
- PMID: 21041489
- PMCID: PMC3019886
- DOI: 10.1128/IAI.00873-10
Quantitative PCR-based competitive index for high-throughput screening of Salmonella virulence factors
Abstract
Salmonella enterica serovar Typhimurium is an intracellular pathogen and a main cause of food-borne illness. In this study, a quantitative PCR (qPCR)-based competitive index (CI) method was developed to simultaneously compare the growth of multiple Salmonella strains. This method was applied to a mixture of 17 Salmonella mutants lacking regulator genes, and their survival ratios were compared based on expression of natural resistance-associated macrophage protein 1 (Nramp1). Nramp1, as a major host innate immune component, controls the intracellular replication of pathogens. Deletion strains containing unique DNA barcodes in place of regulator genes were mixed with the parental control, and the bacteria were inoculated into congenic mice differing only at Nramp1. Most of the deletion strains were outcompeted by wild-type bacteria in either mouse strain, and the lack of Nramp1 didn't increase the tested strain/parent control replication ratios. When the same collection of mutants was tested in congenic mouse-derived primary macrophages, a major Nramp1-expressing cell type, six strains (ΔhimD, ΔphoP/phoQ, ΔrpoE, ΔrpoS, ΔompR/envZ, and Δhfq strains) grew better in Nramp1(-/-) than in Nramp1(+/+) macrophages, suggesting that these six regulators may play roles in overcoming Nramp1-mediated bactericidal activity in primary macrophages. The discrepancy in survival of macrophages and that of mice suggests either that there are differences in macrophage populations or that other cell types expressing Nramp1 control Salmonella proliferation in the host. The method described allows competitive infection analysis to be carried out on complex mixtures of bacteria and provides high reproducibility from independent biological replicates.
Figures
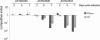
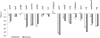
Similar articles
-
Removal of the phage-shock protein PspB causes reduction of virulence in Salmonella enterica serovar Typhimurium independently of NRAMP1.J Med Microbiol. 2014 Jun;63(Pt 6):788-795. doi: 10.1099/jmm.0.072223-0. Epub 2014 Apr 8. J Med Microbiol. 2014. PMID: 24713356
-
Host-pathogen interactions: Host resistance factor Nramp1 up-regulates the expression of Salmonella pathogenicity island-2 virulence genes.Proc Natl Acad Sci U S A. 2002 Nov 26;99(24):15705-10. doi: 10.1073/pnas.252415599. Epub 2002 Nov 19. Proc Natl Acad Sci U S A. 2002. PMID: 12441401 Free PMC article.
-
The Salmonella enterica serovar typhimurium divalent cation transport systems MntH and SitABCD are essential for virulence in an Nramp1G169 murine typhoid model.Infect Immun. 2004 Sep;72(9):5522-5. doi: 10.1128/IAI.72.9.5522-5525.2004. Infect Immun. 2004. PMID: 15322058 Free PMC article.
-
Regulation and function of the Salmonella MgtC virulence protein.J Microbiol. 2015 Oct;53(10):667-72. doi: 10.1007/s12275-015-5283-1. Epub 2015 Aug 1. J Microbiol. 2015. PMID: 26231375 Review.
-
How Pathogens Feel and Overcome Magnesium Limitation When in Host Tissues.Trends Microbiol. 2021 Feb;29(2):98-106. doi: 10.1016/j.tim.2020.07.003. Epub 2020 Aug 14. Trends Microbiol. 2021. PMID: 32807623 Free PMC article. Review.
Cited by
-
Technologies and approaches to elucidate and model the virulence program of salmonella.Front Microbiol. 2011 Jun 2;2:121. doi: 10.3389/fmicb.2011.00121. eCollection 2011. Front Microbiol. 2011. PMID: 21687430 Free PMC article.
-
Evaluation of intracellular survival of Campylobacter fetus subsp. fetus in bovine endometrial cells by qPCR.Iran J Vet Res. 2021 Spring;22(2):94-99. doi: 10.22099/ijvr.2021.38693.5632. Iran J Vet Res. 2021. PMID: 34306105 Free PMC article.
-
A Multi-Omic View of Host-Pathogen-Commensal Interplay in Salmonella-Mediated Intestinal Infection.PLoS One. 2013 Jun 26;8(6):e67155. doi: 10.1371/journal.pone.0067155. Print 2013. PLoS One. 2013. PMID: 23840608 Free PMC article.
-
Global analysis of Salmonella alternative sigma factor E on protein translation.J Proteome Res. 2015 Apr 3;14(4):1716-26. doi: 10.1021/pr5010423. Epub 2015 Feb 27. J Proteome Res. 2015. PMID: 25686268 Free PMC article.
-
ChIP-Seq Analysis of the σE Regulon of Salmonella enterica Serovar Typhimurium Reveals New Genes Implicated in Heat Shock and Oxidative Stress Response.PLoS One. 2015 Sep 21;10(9):e0138466. doi: 10.1371/journal.pone.0138466. eCollection 2015. PLoS One. 2015. PMID: 26389830 Free PMC article.
References
-
- Aberle, S. W., and E. Puchhammer-Stockl. 2002. Diagnosis of herpesvirus infections of the central nervous system. J. Clin. Virol. 25(Suppl. 1):S79-S85. - PubMed
-
- Beuzon, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. - PubMed
-
- Blackwell, J. M., S. Searle, T. Goswami, and E. N. Miller. 2000. Understanding the multiple functions of Nramp1. Microbes Infect. 2:317-321. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

